Safety Assessment of Lactiplantibacillus plantarum GUANKE Based on Whole-Genome Sequencing, Phenotypic, and Anti-Inflammatory Capacity Analysis
- PMID: 40284709
- PMCID: PMC12029457
- DOI: 10.3390/microorganisms13040873
Safety Assessment of Lactiplantibacillus plantarum GUANKE Based on Whole-Genome Sequencing, Phenotypic, and Anti-Inflammatory Capacity Analysis
Abstract
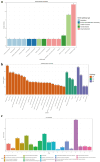
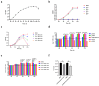
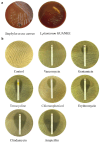
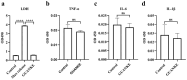
Lactiplantibacillus plantarum GUANKE (L. plantarum GUANKE) is a Gram-positive bacterium isolated from the feces of healthy volunteers. Whole-genome sequencing analysis (WGS) revealed that the genome of L. plantarum GUANKE consists of one chromosome and two plasmids, with the chromosome harbors 2955 CDS, 66 tRNAs, and 5 rRNAs. The genome is devoid of virulence factors and Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems. It contains three intact prophage regions and bacteriocin biosynthesis genes (plantaricins K, F, and E), as well as seventeen genomic islands lacking antibiotic resistance or pathogenicity determinants. Functional prediction outcomes identified that the genome of L. plantarum GUANKE is closely related to transcription, carbohydrate transport and metabolism, and amino acid transport and metabolism. Carbohydrate-active enzymes (CAZymes) analysis and GutSMASH analysis revealed that the genome of L. plantarum GUANKE contained 100 carbohydrate-active enzyme genes and two specialized metabolic gene clusters. Safety assessments confirmed that L. plantarum GUANKE neither exhibited β-hemolytic activity nor harbored detectable transferable drug resistance genes. The strain exhibited remarkable acid tolerance and bile salt resistance. Cellular adhesion assays demonstrated moderate binding capacity to Caco-2 intestinal epithelium (4.3 ± 0.007)%. In vitro analyses using lipopolysaccharide (LPS)-stimulated macrophage models demonstrated that L. plantarum GUANKE significantly suppressed the secretion of pro-inflammatory cytokines (TNF-α, IL-6, IL-1β), exhibiting dose-dependent anti-inflammatory activity. In vivo experiments showed that L. plantarum GUANKE was involved in the regulation of the apical junction pathway and interferon pathway in colon tissue of normal mice.
Keywords: L. plantarum; immunological regulation; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Probiogenomic In-Silico Analysis and Safety Assessment of Lactiplantibacillus plantarum DJF10 Strain Isolated from Korean Raw Milk.Int J Mol Sci. 2022 Nov 21;23(22):14494. doi: 10.3390/ijms232214494. Int J Mol Sci. 2022. PMID: 36430971 Free PMC article.
-
Probiotic Potential and Safety Assessment of Lactiplantibacillus plantarum cqf-43 and Whole-Genome Sequence Analysis.Int J Mol Sci. 2023 Dec 17;24(24):17570. doi: 10.3390/ijms242417570. Int J Mol Sci. 2023. PMID: 38139398 Free PMC article.
-
Administration of Lactobacillus plantarum GUANKE alleviates SARS-CoV-2-induced pneumonia in mice.Microbiol Spectr. 2024 Oct 21;12(12):e0160324. doi: 10.1128/spectrum.01603-24. Online ahead of print. Microbiol Spectr. 2024. PMID: 39431894 Free PMC article.
-
Effects of Lactiplantibacillus plantarum GUANKE on Diphenoxylate-Induced Slow Transit Constipation and Gut Microbiota in Mice.Nutrients. 2023 Aug 26;15(17):3741. doi: 10.3390/nu15173741. Nutrients. 2023. PMID: 37686774 Free PMC article.
-
Whole Genome Analysis of Tibetan Kefir-Derived Lactiplantibacillus Plantarum 12-3 Elucidates Its Genomic Architecture, Antimicrobial and Drug Resistance, Potential Probiotic Functionality and Safety.Front Biosci (Landmark Ed). 2024 Apr 11;29(4):147. doi: 10.31083/j.fbl2904147. Front Biosci (Landmark Ed). 2024. PMID: 38682181
References
-
- Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. Expert Consensus Document. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. - DOI - PubMed
-
- Marteau P. Safety Aspects of Probiotic Products. Näringsforskning. 2001;45:22–24. doi: 10.3402/fnr.v45i0.1785. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

