Comprehensive Genomic Analysis of Klebsiella pneumoniae and Its Temperate N-15-like Phage: From Isolation to Functional Annotation
- PMID: 40284744
- PMCID: PMC12029707
- DOI: 10.3390/microorganisms13040908
Comprehensive Genomic Analysis of Klebsiella pneumoniae and Its Temperate N-15-like Phage: From Isolation to Functional Annotation
Abstract
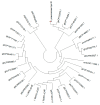
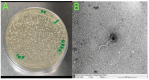
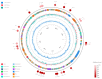
Antibiotic resistance to Klebsiella pneumoniae poses a major public health threat, particularly in intensive care unit (ICU) settings. The emergence of extensively drug-resistant (XDR) strains complicates treatment options, requiring a deeper understanding of their genetic makeup and potential therapeutic targets. This research delineated an extensively drug-resistant (XDR) Klebsiella pneumoniae strain obtained from an ICU patient and telomeric temperate phage derived from hospital effluent. The bacteria showed strong resistance to multiple antibiotics, including penicillin (≥16 μg/mL), ceftriaxone (≥32 μg/mL), and meropenem (≥8 μg/mL), which was caused by SHV-11 beta-lactamase, NDM-1 carbapenemase, and porin mutations (OmpK37, MdtQ). The strain was categorized as K46 and O2a types and carried virulence genes involved in iron acquisition, adhesion, and immune evasion, as well as plasmids (IncHI1B_1_pNDM-MAR, IncFIB) and eleven prophage regions, reflecting its genetic adaptability and resistance dissemination. The 172,025 bp linear genome and 46.3% GC content of the N-15-like phage showed strong genomic similarities to phages of the Sugarlandvirus genus, especially those that infect K. pneumoniae. There were structural proteins (11.8%), DNA replication and repair enzymes (9.3%), and a toxin-antitoxin system (0.4%) encoded by the phage genome. A protelomerase and ParA/B partitioning proteins indicate that the phage is replicating and maintaining itself in a manner similar to the N15 phage, which is renowned for maintaining a linear plasmid prophage throughout lysogeny. Understanding the dynamics of antibiotic resistance and pathogen development requires knowledge of phages like this one, which are known for their temperate nature and their function in altering bacterial virulence and resistance profiles. The regulatory and structural proteins of the phage also provide a model for research into the biology of temperate phages and their effects on microbial communities. The importance of temperate phages in bacterial genomes and their function in the larger framework of microbial ecology and evolution is emphasized in this research.
Keywords: Klebsiella pneumoniae; functional annotation; phage; temperate.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

