The Virus Entry Pathway Determines Sensitivity to the Antiviral Peptide TAT-I24
- PMID: 40284901
- PMCID: PMC12031635
- DOI: 10.3390/v17040458
The Virus Entry Pathway Determines Sensitivity to the Antiviral Peptide TAT-I24
Abstract
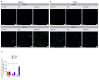
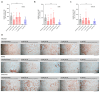
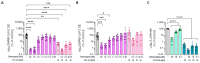
The peptide TAT-I24, a fusion of the TAT peptide (amino acids 48-60) and the 9-mer peptide I24, has been previously shown to neutralize several double-stranded (ds) DNA viruses in vitro. We have now extended the testing to potentially sensitive RNA viruses and analyzed the antiviral effect of the peptide against Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). In Vero E6 cells, TAT-I24 neutralized the human 2019-nCoV isolate (Wuhan variant) in a dose-dependent manner, while it was unable to neutralize two SARS-CoV-2 variants of concern, Delta and Omicron. Moreover, TAT-I24 could not significantly neutralize any of the SARS-CoV-2 variants in the human lung carcinoma cell line Calu-3, which provides an alternative entry route for SARS-CoV-2 by direct membrane fusion. Therefore, a possible dependence on virus uptake by endocytosis was investigated by exposing Vero E6 cells to chloroquine (CQ), an inhibitor of endosomal acidification. The Wuhan variant was highly sensitive to inhibition by CQ, an effect which was further enhanced by TAT-I24, while the Delta variant was less sensitive to inhibition by higher concentrations of CQ compared to the Wuhan variant. The microscopic analysis of COS-7 cells using a rhodamine-labeled TAT-I24 (Rho-TAT-I24) showed the endosomal localization of fluorescent TAT-I24 and co-localization with transfected GFP-Rab14 but not GFP-Rab5. As these proteins are found in distinct endosomal pathways, our results indicate that the virus entry pathway determines sensitivity to the peptide.
Keywords: Rab14; SARS-CoV-2; antiviral peptide; endocytosis; virus entry.
Conflict of interest statement
Hanna Harant owns 100% of the shares of Pivaris BioScience GmbH. Kurt Zatloukal is the co-founder and CEO of Zatloukal-Innovations GmbH. The other authors declare no conflicts of interest.
Figures




References
-
- Lytras S., Xia W., Hughes J., Jiang X., Robertson D.L. The Animal Origin of SARS-CoV-2. Science. 2021;373:968–970. - PubMed
-
- Carabelli A.M., Peacock T.P., Thorne L.G., Harvey W.T., Hughes J., de Silva T.I., Peacock S.J., Barclay W.S., de Silva T.I., Towers G.J., et al. SARS-CoV-2 Variant Biology: Immune Escape, Transmission and Fitness. Nat. Rev. Microbiol. 2023;21:162–177. doi: 10.1038/s41579-022-00841-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

