Expression Profile of Human Cytomegalovirus UL111A cmvIL-10 and LAcmvIL-10 Transcripts in Primary Cells and Cells from Renal Transplant Recipients
- PMID: 40284944
- PMCID: PMC12031159
- DOI: 10.3390/v17040501
Expression Profile of Human Cytomegalovirus UL111A cmvIL-10 and LAcmvIL-10 Transcripts in Primary Cells and Cells from Renal Transplant Recipients
Abstract
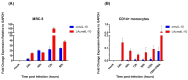
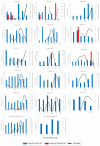
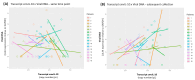
Human cytomegalovirus (HCMV) is a high-risk pathogen in immunocompromised individuals, especially in transplant recipients. HCMV viremia must be monitored, and frequently, patients are treated with antiviral agents. HCMV has a variety of strategies to modulate host antiviral responses, and one important player is a viral homolog of the cellular interleukin-10 (cIL-10). The viral UL111A gene produces several HCMV IL-10 transcripts and protein isoforms through alternative splicing. The cmvIL-10 (isoform A) has similar properties to cIL-10, while LAcmvIL-10 (isoform B) has more restricted biological properties. Other isoforms are produced (C to H) but have unknown functions. Here, we investigated the expression of the most abundant transcripts, cmvIL-10 and LAcmvIL-10, in productively and latently infected cells and in peripheral blood mononuclear cells from renal transplant recipients up to 60 days post-transplantation. This study investigated HCMV cmvIL-10 and LAcmvIL-10 transcription profiles in vitro, in productive and latent infection, and in vivo, in peripheral blood mononuclear cells (PBMCs) of renal transplant patients. In vitro, both cmvIL-10 and LAcmvIL-10 transcripts were detected in both types at high levels and low levels in MRC-5 and latent infected (CD14+). When PBMCs from transplant patients were analyzed, LAcmvIL-10 was detected mostly sporadically and in only a few patients, while cmvIL-10 was found in all patients at all time points. Furthermore, it was observed in PBMCs that expression of cmvIL-10 was positively associated with an increase in viral DNA detection in the subsequently collected sample, indicating that expression of cmvIL-10 might precede viral DNA replication. These results contribute to the understanding of HCMV biology in different phases of infection. In addition, our initial analysis suggests that monitoring cmvIL-10, along with viral DNA, could improve early detection of HCMV reactivation in transplant recipients.
Keywords: HCMV IL-10; HCMV latent infection; HCMV lytic infection; UL111A transcripts; human cytomegalovirus (HCMV); kidney transplant.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Restriction of Human Cytomegalovirus Infection by Galectin-9.J Virol. 2019 Jan 17;93(3):e01746-18. doi: 10.1128/JVI.01746-18. Print 2019 Feb 1. J Virol. 2019. PMID: 30487283 Free PMC article.
-
The effective multiplicity of infection for HCMV depends on the activity of the cellular 20S proteasome.J Virol. 2025 Jan 31;99(1):e0175124. doi: 10.1128/jvi.01751-24. Epub 2024 Dec 10. J Virol. 2025. PMID: 39655950 Free PMC article.
-
Third intracellular loop of HCMV US28 is necessary for signaling and viral reactivation.J Virol. 2025 Jan 31;99(1):e0180124. doi: 10.1128/jvi.01801-24. Epub 2024 Dec 10. J Virol. 2025. PMID: 39655954 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Immunosuppressive T-cell antibody induction for heart transplant recipients.Cochrane Database Syst Rev. 2013 Dec 2;2013(12):CD008842. doi: 10.1002/14651858.CD008842.pub2. Cochrane Database Syst Rev. 2013. PMID: 24297433 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

