Design and Characterization of Inhibitors of Cell-Mediated Degradation of APOBEC3G That Decrease HIV-1 Infectivity
- PMID: 40284957
- PMCID: PMC12031279
- DOI: 10.3390/v17040514
Design and Characterization of Inhibitors of Cell-Mediated Degradation of APOBEC3G That Decrease HIV-1 Infectivity
Abstract
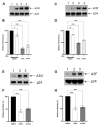
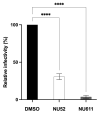
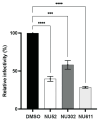
The cytoplasmic human Apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3 or A3) cytidine deaminases G and F (A3G and A3F) can block the spread of human immunodeficiency virus (HIV). HIV counteracts this cell-intrinsic defense through a viral protein called viral infectivity factor (Vif). Vif causes proteasomal degradation of A3G and A3F proteins (A3G/F) in HIV-producing cells to ensure infectivity of virions subsequently released from these cells. Here, we optimized a lead compound reported previously to boost cellular levels of A3G. The modified analogs designed, synthesized, and evaluated here inhibit cell-mediated post-translational degradation of A3G/F, whether Vif is present or not. This increases A3G/F incorporation into Vif-positive virions to decrease viral infectivity. The compounds and processes described here can facilitate the development of new anti-HIV therapeutics whose host-targeted effect may not be evaded by resistance-conferring mutations in HIV Vif.
Keywords: APOBEC3F; APOBEC3G; HIV; HIV cure research; HIV infectivity; HIV-1; Vif.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Conticello S.G., Thomas C.J., Petersen-Mahrt S.K., Neuberger M.S. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005;22:367–377. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

