Molecular Epidemiology of SARS-CoV-2 in Bangladesh
- PMID: 40284960
- PMCID: PMC12031083
- DOI: 10.3390/v17040517
Molecular Epidemiology of SARS-CoV-2 in Bangladesh
Abstract
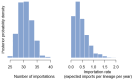
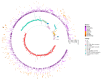
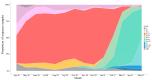
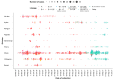
Mutation is one of the most important drivers of viral evolution and genome variability, allowing viruses to potentially evade host immune responses and develop drug resistance. In the context of COVID-19, local genomic surveillance of circulating virus populations is therefore critical. The goals of this study were to describe the distribution of different SARS-CoV-2 lineages, assess their genomic differences, and infer virus importation events in Bangladesh. We individually aligned 1965 SARS-CoV-2 genome sequences obtained between April 2020 and June 2021 to the Wuhan-1 sequence and used the resulting multiple sequence alignment as input to infer a maximum likelihood phylogenetic tree. Sequences were assigned to lineages as described by the hierarchical Pangolin nomenclature scheme. We built a phylogeographic model using the virus population genome sequence variation to infer the number of virus importation events. We observed thirty-four lineages and sub-lineages in Bangladesh, with B.1.1.25 and its sub-lineages D.* (979 sequences) dominating, as well as the Beta variant of concern (VOC) B.1.351 and its sub-lineages B.1.351.* (403 sequences). The earliest B.1.1.25/D.* lineages likely resulted from multiple introductions, some of which led to larger outbreak clusters. There were 570 missense mutations, 426 synonymous mutations, 18 frameshift mutations, 7 deletions, 2 insertions, 10 changes at start/stop codons, and 64 mutations in intergenic or untranslated regions. According to phylogeographic modeling, there were 31 importation events into Bangladesh (95% CI: 27-36). Like elsewhere, Bangladesh has experienced distinct waves of dominant lineages during the COVID-19 pandemic; this study focuses on the emergence and displacement of the first wave-dominated lineage, which contains mutations seen in several VOCs and may have had a transmission advantage over the extant lineages.
Keywords: Bangladesh; SARS-CoV-2; epidemiology; mutation; phylogeny; spike protein.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- WHO . WHO Bangladesh COVID-19 Morbidity and Mortality Weekly Update (MMWU) Volume 70 WHO; Geneva, Switzerland: 2021.
-
- Mohmmad Mahmud A.S., Taznin T., Hasan Sarkar M.M., Uzzaman M.S., Osman E., Habib M.A., Akter S., Banu T.A., Goswami B., Jahan I., et al. The genetic variant analyses of SARS-CoV-2 strains; circulating in Bangladesh. bioRxiv. 2020 doi: 10.1101/2020.07.29.226555. - DOI
-
- Parvin R., Afrin S.Z., Begum J.A., Ahmed S., Nooruzzaman M., Chowdhury E.H., Pohlmann A., Paul S.K. Molecular Analysis of SARS-CoV-2 Circulating in Bangladesh during 2020 Revealed Lineage Diversity and Potential Mutations. Microorganisms. 2021;9:1035. doi: 10.3390/microorganisms9051035. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

