The Ongoing Epidemics of Seasonal Influenza A(H3N2) in Hangzhou, China, and Its Viral Genetic Diversity
- PMID: 40284969
- PMCID: PMC12031544
- DOI: 10.3390/v17040526
The Ongoing Epidemics of Seasonal Influenza A(H3N2) in Hangzhou, China, and Its Viral Genetic Diversity
Abstract
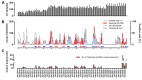
This study examined the genetic and evolutionary features of influenza A/H3N2 viruses in Hangzhou (2010-2022) by analyzing 28,651 influenza-like illness samples from two sentinel hospitals. Influenza A/H3N2 coexisted with other subtypes, dominating seasonal peaks (notably summer). Whole-genome sequencing of 367 strains was performed on GridION platforms. Phylogenetic analysis showed they fell into 16 genetic groups, with multiple clades circulating simultaneously. Shannon entropy indicated HA, NA, and NS gene segments exhibited significantly higher variability than other genomic segments, with HA glycoprotein mutations concentrated in antigenic epitopes A-E. Antiviral resistance showed no inhibitor resistance mutations in PA, PB1, or PB2, but NA mutations were detected in some strains, and most strains harbored M2 mutations. A Bayesian molecular clock showed the HA segment exhibited the highest nucleotide substitution rate (3.96 × 10-3 substitutions/site/year), followed by NA (3.77 × 10-3) and NS (3.65 × 10-3). Selective pressure showed A/H3N2 strains were predominantly under purifying selection, with only sporadic positive selection at specific sites. The Pepitope model demonstrated that antigenic epitope mismatches between circulating H3N2 variants and vaccine strains led to a significant decline in influenza vaccine effectiveness (VE), particularly in 2022. Overall, the study underscores the complex circulation patterns of influenza in Hangzhou and the global importance of timely vaccine strain updates.
Keywords: Bayesian molecular clock; Pepitope model; Shannon entropy; antiviral resistance; influenza A/H3N2; phylogenetic analysis; selective pressure; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Jallow M.M., Barry M.A., Ndiaye N.K., Touré C.T., Kiori D., Sagne S.N., Sy S., Goudiaby D., Niang M.N., Diagne M.M., et al. Genetic and antigenic characterization of influenza A(H3N2) virus after 13 consecutive years of influenza surveillance in Senegal, 2010–2022. J. Med. Virol. 2024;96:e70010. doi: 10.1002/jmv.70010. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2023RC072/Medicine and Health Technology Project of Zhejiang Province
- 31301098/National Natural Science Foundation of China
- LGF19H190003/Zhejiang Provincial Public Welfare Technology Application Research Project of China
- 2024ZY01026/Zhejiang Provincial Key Laboratory Construction Project
- 2018-2022/Construction Fund of Key Medical Disciplines of Hangzhou and the Zhejiang Provincial Pro-gramme for the Cultivation of High-level Innovative Health Talents
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

