Genomic and Epidemiological Investigations Reveal Chromosomal Integration of the Acipenserid Herpesvirus 3 Genome in Lake Sturgeon Acipenser fulvescens
- PMID: 40284977
- PMCID: PMC12031113
- DOI: 10.3390/v17040534
Genomic and Epidemiological Investigations Reveal Chromosomal Integration of the Acipenserid Herpesvirus 3 Genome in Lake Sturgeon Acipenser fulvescens
Abstract
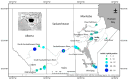
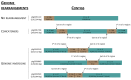
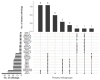
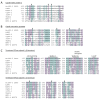
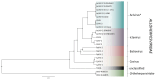
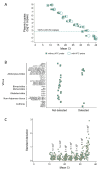
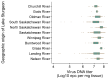
DNA sequence from a new alloherpesvirus named acipenserid herpesvirus 3 (AciHV-3) was found in sturgeon species that are vulnerable to decline globally. A study was undertaken to develop a better understanding of the virus genome and to develop diagnostic tools to support an epidemiological investigation. A 184,426 bp genome was assembled from PacBio HiFi sequences generated with DNA from a Lake Sturgeon Acipenser fulvescens gonad cell line. The AciHV-3 genome was contiguous with host chromosomal DNA and was structured with telomere-like terminal direct repeat regions, five internal direct repeat regions and a U region that included intact open reading frames encoding alloherpesvirus core proteins. Diagnostic testing conducted with a newly developed and analytically validated qPCR assay established the ubiquitous presence and high titer of AciHV-3 DNA in somatic and germline tissues from wild Lake Sturgeon in the Hudson Bay drainage basin. Phylogenetic reconstructions confirm that the monophyletic AciHV-3 lineage shares a common ancestor with AciHV-1 and that AciHV-3 taxa cluster according to their sturgeon host. The same genotype of AciHV-3 is found in disjunctive Lake Sturgeon populations within and among drainage basins. The results support the hypotheses that AciHV-3 has established latency through germline chromosomal integration, is vertically transmitted via a Mendelian pattern of inheritance, is evolving in a manner consistent with a replication competent virus and has co-evolved with its host reaching genetic fixation in Lake Sturgeon populations in central Canada.
Keywords: Alloherpesviridae; Lake Sturgeon; endogenous; germline; herpesvirus.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


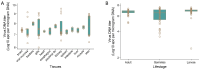

Similar articles
-
Partial genome analysis of Siberian sturgeon alloherpesvirus suggests its close relation to AciHV-2 - short communication.Acta Vet Hung. 2010 Jun;58(2):269-74. doi: 10.1556/AVet.58.2010.2.13. Acta Vet Hung. 2010. PMID: 20460226
-
Molecular detection and characterisation of herpesviruses in asymptomatic Russian sturgeon (Acipenser gueldenstaedtii) from European aquaculture.J Vet Res. 2025 May 21;69(2):169-175. doi: 10.2478/jvetres-2025-0028. eCollection 2025 Jun. J Vet Res. 2025. PMID: 40552023 Free PMC article.
-
First report of ictavirus acipenseridallo2 (AciHV-2) in Danube sturgeons Acipenser gueldenstaedtii in Austria.Dis Aquat Organ. 2025 Feb 6;161:47-53. doi: 10.3354/dao03837. Dis Aquat Organ. 2025. PMID: 39912416
-
NIH Consensus Statement on Management of Hepatitis C: 2002.NIH Consens State Sci Statements. 2002 Jun 10-12;19(3):1-46. NIH Consens State Sci Statements. 2002. PMID: 14768714
-
Lysine supplementation is not effective for the prevention or treatment of feline herpesvirus 1 infection in cats: a systematic review.BMC Vet Res. 2015 Nov 16;11:284. doi: 10.1186/s12917-015-0594-3. BMC Vet Res. 2015. PMID: 26573523 Free PMC article.
References
-
- ICTV 2024 (International Committee on Taxonomy of Viruses). 2023 Taxonomy Release. [(accessed on 20 September 2024)]. Available online: https://ictv.global/taxonomy.
-
- McGeoch D.J., Rixon F.J., Davison A.J. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117:90–104. - PubMed
-
- McGeoch D.J., Davison A.J., Dolan A., Gatherer D., Sevilla-Reyes E.E. Molecular evolution of the Herpesvirales. In: Domingo E., Parrish C.R., Holland J.J., editors. Origin and Evolution of Viruses. 2nd ed. Academic Press; London, UK: 2008. pp. 447–475.
-
- Roizman B., Pellett P.E. The family of Herpesviridae: A brief introduction. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2001. pp. 2381–2397.
-
- Roizman B., Knipe D.M., Whitley R.J. Herpes simplex viruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 2501–2601.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

