Phage Endolysins as Promising and Effective Candidates for Use Against Uropathogenic Escherichia coli
- PMID: 40285003
- PMCID: PMC12031403
- DOI: 10.3390/v17040560
Phage Endolysins as Promising and Effective Candidates for Use Against Uropathogenic Escherichia coli
Abstract
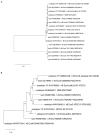
The presented in silico and phylogenetic analysis of putative endolysins potentially produced by phages infecting uropathogenic Escherichia coli (UPEC) demonstrates their remarkable diversity. These proteins exhibit significant variations in sequence length, molecular weight, isoelectric point, and stability, as well as diverse functional domains determining their enzymatic activity, including lysin, lysozyme, hydrolase, amidase, and peptidase functions. Due to their predicted lytic properties, endolysins hold great promise in combating UPEC bacteria, including those within biofilms, which are often highly resistant to conventional treatments. Despite their potential, several challenges hinder the full utilization of endolysins. These include the relatively small number of identified proteins, challenges in the annotation process, and the scarcity of studies evaluating their efficacy in vitro and in vivo against Gram-negative bacteria. In this work, we emphasize these challenges while also underlining the potential of endolysins as an effective tool against UPEC infections. Their effectiveness could be significantly enhanced when combined with agents that disrupt the outer membrane of these bacteria, making them a promising alternative or complement to existing antimicrobial strategies. Further research is necessary to fully explore their therapeutic potential.
Keywords: bacteriophages; phage endolysins; phage therapy; urinary tract infections (UTIs); uropathogenic Escherichia coli (UPEC).
Conflict of interest statement
Author Sylwia Bloch and Bożena Nejman-Faleńczyk were employed by the company BNF—New Bio Force Ltd., The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

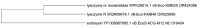








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

