Expression Profiles of lncRNAs and mRNAs in the Mouse Brain Infected with Pseudorabies Virus: A Bioinformatic Analysis
- PMID: 40285022
- PMCID: PMC12031243
- DOI: 10.3390/v17040580
Expression Profiles of lncRNAs and mRNAs in the Mouse Brain Infected with Pseudorabies Virus: A Bioinformatic Analysis
Abstract
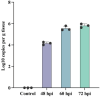
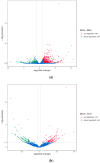
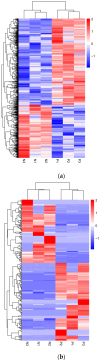
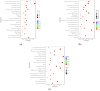
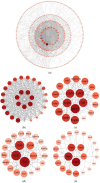
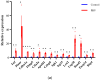
Pseudorabies virus (PRV) is a pathogen that causes severe neurological dysfunction in the host, leading to extensive neuronal damage and inflammation. Despite extensive research on the neuropathogenesis of α-herpesvirus infections, many scientific questions remain unresolved, such as the largely unknown functions of long non-coding RNAs (lncRNAs) in herpesvirus-infected nervous systems. To address these questions, we used RNA sequencing (RNA-seq) to investigate the expression profiles of lncRNAs and mRNAs in the brains of mice infected with PRV. Through bioinformatic analysis, we identified 316 differentially expressed lncRNAs and 886 differentially expressed mRNAs. We predicted the biological functions of these differentially expressed lncRNAs and mRNAs using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, and the results showed that the differentially expressed transcripts were mainly involved in the innate immune response. Finally, we validated the differential expression trends of lncRNAs and mRNAs using quantitative real-time PCR (q-PCR), which were consistent with the sequencing data. To our knowledge, this is the first report analyzing the lncRNA expression profile in the central nervous system (CNS) of mice infected with PRV. Our findings provide new insights into the roles of lncRNAs and mRNAs during PRV infection of the host CNS.
Keywords: RNA-seq; brain; lncRNA; pseudorabies virus.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Dysregulated lncRNAs regulate human umbilical cord mesenchymal stem cell differentiation into insulin-producing cells by forming a regulatory network with mRNAs.Stem Cell Res Ther. 2024 Jan 25;15(1):22. doi: 10.1186/s13287-023-03572-5. Stem Cell Res Ther. 2024. PMID: 38273351 Free PMC article.
-
Transcriptomic profiling and bioinformatic insights into myocardial injury following aneurysmal subarachnoid hemorrhage.Front Neurol. 2025 Jun 24;16:1492398. doi: 10.3389/fneur.2025.1492398. eCollection 2025. Front Neurol. 2025. PMID: 40630915 Free PMC article.
-
Identification of Key Genes for Alcoholic Hepatitis Using Integrated Network Analysis of Differential lncRNA and Gene Expression.Int J Mol Sci. 2025 Jun 25;26(13):6104. doi: 10.3390/ijms26136104. Int J Mol Sci. 2025. PMID: 40649881 Free PMC article.
-
Role of long non-coding RNAs in neurofibromatosis and Schwannomatosis: pathogenesis and therapeutic potential.Epigenomics. 2024 Dec-Dec;16(23-24):1453-1464. doi: 10.1080/17501911.2024.2430170. Epub 2024 Nov 27. Epigenomics. 2024. PMID: 39601046 Review.
-
The Expression and Regulatory Roles of Long Non-Coding RNAs in Periodontal Ligament Cells: A Systematic Review.Biomolecules. 2022 Feb 12;12(2):304. doi: 10.3390/biom12020304. Biomolecules. 2022. PMID: 35204802 Free PMC article.
References
-
- Papageorgiou K., Stoikou A., Papadopoulos D.K., Tsapouri-Kanoula E., Giantsis I.A., Papadopoulos D., Stamelou E., Sofia M., Billinis C., Karapetsiou C., et al. Pseudorabies Virus Prevalence in Lung Samples of Hunted Wild Boars in Northwestern Greece. Pathogens. 2024;13:929. doi: 10.3390/pathogens13110929. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

