Fecal Microbiome Reflects Disease State and Prognosis in Inflammatory Bowel Disease in an Adult Population-Based Inception Cohort
- PMID: 40285477
- PMCID: PMC12491950
- DOI: 10.1093/ibd/izaf060
Fecal Microbiome Reflects Disease State and Prognosis in Inflammatory Bowel Disease in an Adult Population-Based Inception Cohort
Abstract
Introduction: We aimed to determine the diagnostic and prognostic potential of baseline microbiome profiling in inflammatory bowel disease (IBD).
Methods: Participants with ulcerative colitis (UC), Crohn's disease (CD), suspected IBD, and non-IBD symptomatic controls were included in the prospective population-based cohort Inflammatory Bowel Disease in South-Eastern Norway III (third iteration) based on suspicion of IBD. The participants donated fecal samples that were analyzed with 16S rRNA sequencing. Disease course severity was evaluated at the 1-year follow-up. A stringent statistical consensus approach for differential abundance analysis with 3 different tools was applied, together with machine learning modeling.
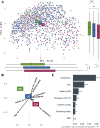
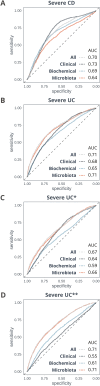
Results: A total of 1404 individuals were included, where n = 1229 samples from adults were used in the main analyses (n = 658 UC, n = 324 CD, n = 36 IBD-U, n = 67 suspected IBD, and n = 144 non-IBD symptomatic controls). Microbiome profiles were compared with biochemical markers in machine learning models to differentiate IBD from non-IBD symptomatic controls (area under the receiver operating curve [AUC] 0.75-0.79). For UC vs controls, integrating microbiome data with biochemical markers like fecal calprotectin mildly improved classification (AUC 0.83 to 0.86, P < .0001). Extensive differences in microbiome composition between UC and CD were identified, which could be quantified as an index of differentially abundant genera. This index was validated across published datasets from 3 continents. The UC-CD index discriminated between ileal and colonic CD (linear regression, P = .008) and between colonic CD and UC (P = .005), suggesting a location-dependent gradient. Microbiome profiles outperformed biochemical markers in predicting a severe disease course in UC (AUC 0.72 vs 0.65, P < .0001), even in those with a mild disease at baseline (AUC 0.66 vs 0.59, P < .0001).
Conclusions: Fecal microbiome profiling at baseline held limited potential to diagnose IBD from non-IBD compared with standard-of-care. However, microbiome shows promise for predicting future disease courses in UC.
Keywords: Crohn’s disease; IBSEN; machine learning; ulcerative colitis.
Plain language summary
This study assessed the diagnostic and prognostic potential of baseline microbiome profiling in IBD. The microbiome holds the potential to differentiate IBD from controls and enhanced predictive capacity for UC severity. A UC-CD microbial index distinguished disease types and was validated with a global cohort.
© 2025 Crohn’s & Colitis Foundation. Published by Oxford University Press on behalf of Crohn’s & Colitis Foundation.
Conflict of interest statement
M.G.M. holds shares in Bio-Me AS. S.O.F. reports personal fees during the last 2 years from Takeda, Galapagos, Jansen-Cilag, AbbVie, Pharmacosmos, Norgine, and Bristol-Myers-Squibb. The remaining authors disclose no conflicts.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

