Pushing the Frontiers: Artificial Intelligence (AI)-Guided Programmable Concepts in Binary Self-Assembly of Colloidal Nanoparticles
- PMID: 40285639
- PMCID: PMC12302557
- DOI: 10.1002/advs.202501000
Pushing the Frontiers: Artificial Intelligence (AI)-Guided Programmable Concepts in Binary Self-Assembly of Colloidal Nanoparticles
Abstract
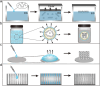
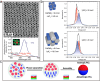
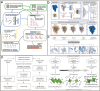
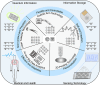
Colloidal nanoparticle self-assembly is a key area in nanomaterials science, renowned for its ability to design metamaterials with tailored functionalities through a bottom-up approach. Over the past three decades, advancements in nanoparticle synthesis and assembly control methods have propelled the transition from single-component to binary assemblies. While binary assembly has been recognized as a significant concept in materials design, its potential for intelligent and customized assembly has often been overlooked. It is argued that the future trend in the assembly of binary nanocrystalline superlattices (BNLSs) can be analogous to the '0s' and '1s' in computer programming, and customizing their assembly through precise control of these basic units could significantly expand their application scope. This review briefly recaps the developmental trajectory of nanoparticle assembly, tracing its evolution from simple single-component assemblies to complex binary co-assemblies and the unique property changes they induce. Of particular significance, this review explores the future prospects of binary co-assembly, viewed through the lens of 'AI-guided programmable assembly'. Such an approach has the potential to shift the paradigm from passive assembly to active, intelligent design, leading to the creation of new materials with disruptive properties and functionalities and driving profound changes across multiple high-tech fields.
Keywords: artificial intelligence; binary co‐assembly; colloidal nanoparticles; intelligent novel materials; self‐assembly; superstructural materials.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures











References
-
- a) Xue Z., Yan C., Wang T., Adv. Funct. Mater. 2019, 29, 1807658;
- b) Boles M. A., Engel M., Talapin D. V., Chem. Rev. 2016, 116, 11220; - PubMed
- c) Wang T., Zhuang J., Lynch J., Chen O., Wang Z., Wang X., LaMontagne D., Wu H., Wang Z., Cao Y. C., Science 2012, 338, 358; - PubMed
- d) Nagaoka Y., Tan R., Li R., Zhu H., Eggert D., Wu Y. A., Liu Y., Wang Z., Chen O., Nature 2018, 561, 378; - PubMed
- e) Nagaoka Y., Zhu H., Eggert D., Chen O., Science 2018, 362, 1396; - PubMed
- f) Chen O., Riedemann L., Etoc F., Herrmann H., Coppey M., Barch M., Farrar C. T., Zhao J., Bruns O. T., Wei H., Guo P., Cui J., Jensen R., Chen Y., Harris D. K., Cordero J. M., Wang Z., Jasanoff A., Fukumura D., Reimer R., Dahan M., Jain R. K., Bawendi M. G., Nat. Commun. 2014, 5, 5093; - PMC - PubMed
- g) Castelli A., de Graaf J., Marras S., Brescia R., Goldoni L., Manna L., Arciniegas M. P., Nat. Commun. 2018, 9, 1141; - PMC - PubMed
- h) Wang X., Zhuang J., Peng Q., Li Y., Nature 2005, 437, 121; - PubMed
- i) Bo A., Liu Y., Kuttich B., Kraus T., Widmer‐Cooper A., de Jonge N., Adv. Mater. 2022, 34, 2109093; - PubMed
- j) Qiao X., Su B., Liu C., Song Q., Luo D., Mo G., Wang T., Adv. Mater. 2018, 30, 1702275; - PubMed
- k) Huang H., Li J., Yuan M., Yang H., Zhao Y., Ying Y., Wang S., Angew. Chem. Int. Ed. 2022, 61, 202211163; - PubMed
- l) Prasad B. L. V., Sorensen C. M., Klabunde K. J., superlattices G., Chem. Soc. Rev. 2008, 37, 1871; - PubMed
- m) Wang D., Xie T., Peng Q., Li Y., J. Am. Chem. Soc. 2008, 130, 4016 ; - PubMed
- n) Leng K., Wang L., Shao Y., Abdelwahab I., Grinblat G., Verzhbitskiy I., Li R., Cai Y., Chi X., Fu W., Song P., Rusydi A., Eda G., Maier S. A., Loh K. P., Nat. Commun. 2020, 11, 5483; - PMC - PubMed
- o) Jana A., Meena A., Patil S. A., Jo Y., Cho S., Park Y., Sree V. G., Kim H., Im H., Taylor R. A., Prog. Mater. Sci. 2022, 129, 100975;
- p) Tong Y., Yao E. P., Manzi A., Bladt E., Wang K., Doblinger M., Bals S., Muller‐Buschbaum P., Urban A. S., Polavarapu L., Feldmann J., Adv. Mater. 2018, 30, 1801117; - PubMed
- q) Mosquera J., Wang D., Bals S., Liz‐Marzán L. M., Nanorods S. L. G., Acc. Chem. Res. 2023, 56, 1204; - PMC - PubMed
- r) Chen O., Zhao J., Chauhan V. P., Cui J., Wong C., Harris D. K., Wei H., Han H.‐S., Fukumura D., Jain R. K., Bawendi M. G., Nat. Mater. 2013, 12, 445. - PMC - PubMed
-
- a) Zhang N.‐N., Mychinko M., Gao S.‐Y., Yu L., Shen Z.‐L., Wang L., Peng F., Wei Z., Wang Z., Zhang W., Zhu S., Yang Y., Sun T., Liz‐Marzán L. M., Bals S., Liu K., Nano Lett. 2024, 24, 13027; - PubMed
- b) Ni B., Mychinko M., Gómez‐Graña S., Morales‐Vidal J., Obelleiro‐Liz M., Heyvaert W., Vila‐Liarte D., Zhuo X., Albrecht W., Zheng G., González‐Rubio G., Taboada J. M., Obelleiro F., López N., Pérez‐Juste J., Pastoriza‐Santos I., Cölfen H., Bals S., Liz‐Marzán L. M., Adv. Mater. 2023, 35, 2208299; - PubMed
- c) Ni B., González‐Rubio G., Van Gordon K., Bals S., Kotov N. A., Liz‐Marzán L. M., Adv. Mater. 2024, 36, 2412473; - PMC - PubMed
- d) González‐Rubio G., Mosquera J., Kumar V., Pedrazo‐Tardajos A., Llombart P., Solís D. M., Lobato I., Noya E. G., Guerrero‐Martínez A., Taboada J. M., Obelleiro F., MacDowell L. G., Bals S., Liz‐Marzán L. M., Micelle‐dir. chiral seeded growth anisotropic gold nanocrystals, Science 2020, 368, 1472; - PubMed
- e) Van Gordon K., Girod R., Bevilacqua F., Bals S., Liz‐Marzán L. M., Nano Lett. 2025, 25, 2887. - PubMed
-
- a) Bladt E., van Dijk‐Moes R. J. A., Peters J., Montanarella F., de Mello Donega C., Vanmaekelbergh D., Bals S., J. Am. Chem. Soc. 2016, 138, 14288; - PubMed
- b) Sánchez‐Iglesias A., Winckelmans N., Altantzis T., Bals S., Grzelczak M., Liz‐Marzán L. M., J. Am. Chem. Soc. 2017, 139, 107; - PubMed
- c) Grzelczak M., Pérez‐Juste J., Mulvaney P., Liz‐Marzán L. M., Chem. Soc. Rev. 2008, 37, 1783; - PubMed
- d) Li Y. H., Zhao S. N., Zang S. Q., Exploration 2023, 3, 20220005. - PMC - PubMed
-
- a) Carbone L., Nobile C., De Giorgi M., Sala F. D., Morello G., Pompa P., Hytch M., Snoeck E., Fiore A., Franchini I. R., Nadasan M., Silvestre A. F., Chiodo L., Kudera S., Cingolani R., Krahne R., Manna L., Nano Lett. 2007, 7, 2942; - PubMed
- b) Murray C. B., Norris D. J., Bawendi M. G., J. Am. Chem. Soc. 1993, 115, 8706;
- c) Ha J. M., Lim S. H., Dey J., Lee S. J., Lee M. J., Kang S. H., Jin K. S., Choi S. M., Nano Lett. 2019, 19, 2313; - PubMed
- d) Liu Z., Pascazio R., Goldoni L., Maggioni D., Zhu D., Ivanov Y. P., Divitini G., Camarelles J. L., Jalali H. B., Infante I., De Trizio L., Manna L., J. Am. Chem. Soc. 2023, 145, 18329. - PMC - PubMed
-
- a) Kotov N. A., Meldrum F. C., Wu C., Fendler J. H., J. Phys. Chem. 1994, 98, 2735;
- b) Huang C., Chen X., Xue Z., Wang T., Sci. Adv. 2020, 6, aba1321; - PMC - PubMed
- c) Liu J., Huang J., Niu W., Tan C., Zhang H., Chem. Rev. 2021, 121, 5830; - PubMed
- d) Zhuang J., Wu H., Yang Y., Cao Y. C., J. Am. Chem. Soc. 2007, 129, 14166. - PubMed
Publication types
Grants and funding
- 2023YFB3210100/Key Technologies Research and Development Program
- 21925405/National Outstanding Youth Science Fund Project of National Natural Science Foundation of China
- 22304129/Youth Fund of the National Natural Science Foundation of China
- 23JCQNJC00810/Natural Science Foundation of Tianjin
- 24JCZDJC00710/Natural Science Foundation of Tianjin
LinkOut - more resources
Full Text Sources
Miscellaneous
