Insights into Siglec-7 Binding to Gangliosides: NMR Protein Assignment and the Impact of Ligand Flexibility
- PMID: 40285643
- PMCID: PMC12140324
- DOI: 10.1002/advs.202415782
Insights into Siglec-7 Binding to Gangliosides: NMR Protein Assignment and the Impact of Ligand Flexibility
Abstract
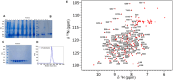
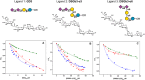
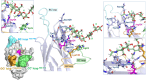
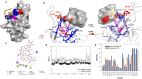
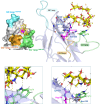
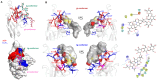
Gangliosides, sialylated glycosphingolipids abundant in the nervous system, play crucial roles in neurotransmission, interaction with regulatory proteins, cell-cell recognition, and signaling. Altered gangliosides expression has been correlated with pathological processes, including cancer, inflammatory disorders, and autoimmune diseases. Gangliosides are important endogenous ligands of Siglecs (Sialic acid-binding immunoglobulin-type lectins), I-type lectins mostly expressed by immune cells, that specifically recognize sialylated glycans. Siglec-7, an inhibitory immune receptor on human natural killer cells, represents a potential target for tumor immunotherapy. Notably, the expression of Siglec-7 ligands is high in various cancers, such as pancreatic cancer and melanoma and lead to tumor immune evasion. Siglec-7 binds the disialylated ganglioside GD3, a tumor-associated antigen overexpressed on cancer cells to suppress immune responses. Using a combination of structural biology techniques, including Nuclear Magnetic Resonance (NMR), biophysical, and computational methods, the binding of Siglec-7 to GD3 and Gb3 derivatives is investigated, revealing the importance of ligand conformation in modulating binding energetics and affinity. The greater flexibility of Gb3 derivatives appears to negatively impact binding entropy, leading to lower affinity compared to GD3. A thorough understanding of these interactions could contribute to elucidating molecular mechanisms of cancer immune evasion and facilitate the development of ganglioside-based diagnostic and therapeutic strategies for cancer.
Keywords: NMR; gangliosides; siglec‐7; structural biology.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Di Carluccio C., Forgione R. E., Molinaro A., Crocker P. R., Marchetti R., Silipo A., Carbohydr. Chem. 2021, 44, 31.
-
- Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L., Seeberger P. H., Essentials of Glycobiology, 4th ed., Cold Spring Harbor Laboratory Press, New York: 2017. - PubMed
-
- Groux‐Degroote S., Guérardel Y., Julien S., Delannoy P., Biochemistry (Moscow) 2015, 80, 808. - PubMed
MeSH terms
Substances
Grants and funding
- 2022ZEZS45/PRIN MUR 2022
- P2022M457Z/PRIN MUR PNRR 2022
- European Research Council (ERC) under the European Union's Horizon 2020 ERC-STG No 851356
- NextGenerationEU, ITACA.SB, Project n° IR0000009), MUR 3264/2021 PNRR M4/C2/L3.1.1
- ProgettoDipartimentidiEccellenza2023-2027toUNINAandUNIFI/Ministry of Education, Universities and Research (Italy)
- PNRR, Missione 4 - Componente 2 - NextGenerationEU - Partenariato Esteso INF-ACT - One Health Basic and Translational Research Actions Addressing Unmet Needs on Emerging Infectious Diseases MUR: PE00000007
- H2020-MSCA-ITN-2020(contractn°956758)/H2020-MSCA-ITN
- Accademia Nazionale dei Lincei
- 25-18490S/ECzech Science Foundation
- LTC20078/Ministry of Youth, Education and Sports of the Czech Republic
- LUAUS25250/Ministry of Youth, Education and Sports of the Czech Republic
- CA18103INNOGLY(STSM)/COST ACTION
LinkOut - more resources
Full Text Sources
Research Materials
