Antiviral Activity of Halogenated Compounds Derived from L-Tyrosine Against SARS-CoV-2
- PMID: 40286029
- PMCID: PMC11990460
- DOI: 10.3390/molecules30071419
Antiviral Activity of Halogenated Compounds Derived from L-Tyrosine Against SARS-CoV-2
Abstract
Introduction: Currently, there are no effective medications for treating all the clinical conditions of patients with COVID-19. We aimed to evaluate the antiviral activity of compounds derived from L-tyrosine against the B.1 lineage of SARS-CoV-2 in vitro and in silico.
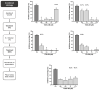
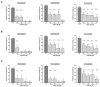
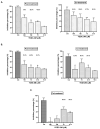
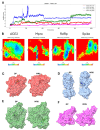
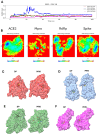
Methodology: The cytotoxicities of 15 halogenated compounds derived from L-tyrosine were evaluated in Vero-E6 cells by the MTT assay. The antiviral activity of the compounds was evaluated using four strategies, and viral quantification was performed by a plaque assay and qRT-PCR. The toxicity of the compounds was evaluated by ADMET predictor software. The affinity of these compounds for viral or cellular proteins and the stability of their conformations were determined by docking and molecular dynamics, respectively.
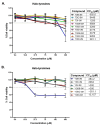
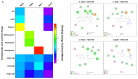
Results: TODC-3M, TODI-2M, and YODC-3M reduced the viral titer >40% and inhibited the replication of viral RNA without significant cytotoxicity. In silico analyses revealed that these compounds presented low toxicity and binding energies between -4.3 and -5.2 Kcal/mol for three viral proteins (spike, Mpro, and RdRp). TODC-3M and YODC-3M presented the most stable conformations with the evaluated proteins.
Conclusions: The most promising compounds were TODC-3M, TODI-2M, and YODC-3M, which presented low in vitro and in silico toxicity, antiviral potential through different strategies, and favorable affinities for viral targets. Therefore, they are candidates for in vivo studies.
Keywords: COVID-19; L-tyrosine; SARS-CoV-2; antiviral activity; halogenated compounds.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
In-vitro and in-silico evaluation of rue herb for SARS-CoV-2 treatment.Int Immunopharmacol. 2024 Dec 25;143(Pt 1):113318. doi: 10.1016/j.intimp.2024.113318. Epub 2024 Oct 10. Int Immunopharmacol. 2024. PMID: 39393270
-
Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2-ACE2 Receptor Interactions.mBio. 2021 Mar 30;12(2):e03681-20. doi: 10.1128/mBio.03681-20. mBio. 2021. PMID: 33785634 Free PMC article.
-
Comprehensive Virtual Screening of the Antiviral Potentialities of Marine Polycyclic Guanidine Alkaloids against SARS-CoV-2 (COVID-19).Biomolecules. 2021 Mar 19;11(3):460. doi: 10.3390/biom11030460. Biomolecules. 2021. PMID: 33808721 Free PMC article.
-
Propolis, Bee Honey, and Their Components Protect against Coronavirus Disease 2019 (COVID-19): A Review of In Silico, In Vitro, and Clinical Studies.Molecules. 2021 Feb 25;26(5):1232. doi: 10.3390/molecules26051232. Molecules. 2021. PMID: 33669054 Free PMC article. Review.
-
[Activity of flavonoids of natural origin on SARS-CoV-2 infections].Postepy Biochem. 2024 Dec 2;70(4):483-489. doi: 10.18388/pb.2021_562. Print 2024 Dec 31. Postepy Biochem. 2024. PMID: 39772327 Review. Polish.
References
-
- WHO COVID-19: Cronología de la Actuación de la OMS. [(accessed on 30 January 2024)];2020 Available online: https://www.who.int/es/news/item/27-04-2020-who-timeline---covid-19.
-
- WHO COVID-19 Epidemiological Update. [(accessed on 30 January 2024)];2024 Available online: https://www.who.int/publications/m/item/covid-19-epidemiological-update-....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

