Protein O-Fucosyltransferases: Biological Functions and Molecular Mechanisms in Mammals
- PMID: 40286076
- PMCID: PMC11990869
- DOI: 10.3390/molecules30071470
Protein O-Fucosyltransferases: Biological Functions and Molecular Mechanisms in Mammals
Abstract
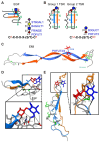
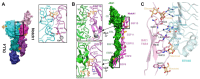
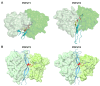
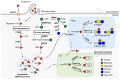
Domain-specific O-fucosylation is an unusual type of glycosylation, where the fucose is directly attached to the serine or threonine residues in specific protein domains via an O-linkage. O-fucosylated proteins play critical roles in a wide variety of biological events and hold important therapeutic values, with the most studied being the Notch receptors and ADAMTS proteins. O-fucose glycans modulate the function of the proteins they modify and are closely associated with various diseases including cancer. In mammals, alongside the well-documented protein O-fucosyltransferase (POFUT) 1-mediated O-fucosylation of epidermal growth factor-like (EGF) repeats and POFUT2-mediated O-fucosylation of thrombospondin type 1 repeats (TSRs), a new type of O-fucosylation was recently identified on elastin microfibril interface (EMI) domains, mediated by POFUT3 and POFUT4 (formerly FUT10 and FUT11). In this review, we present an overview of our current knowledge of O-fucosylation, integrating the latest findings and with a particular focus on its biological functions and molecular mechanisms.
Keywords: EGF; EMI; FUT10; FUT11; Notch; O-fucosylation; POFUT1; POFUT2; POFUT3; POFUT4; TSR.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Ginsburg V. Formation of guanosine diphosphate L-fucose from guanosine diphosphate D-mannose. J. Biol. Chem. 1960;235:2196–2201. - PubMed
-
- Tonetti M., Sturla L., Bisso A., Benatti U., De Flora A. Synthesis of GDP-L-fucose by the human FX protein. J. Biol. Chem. 1996;271:27274–27279. - PubMed
-
- Coffey J.W., Miller O.N., Sellinger O.Z. The metabolism of L-fucose in the rat. J. Biol. Chem. 1964;239:4011–4017. - PubMed
-
- Kaufman R.L., Ginsburg V. The metabolism of L-fucose by HeLa cells. Exp. Cell Res. 1968;50:127–132. - PubMed
-
- Ishihara H., Heath E.C. The metabolism of L-fucose. IV. The biosynthesis of guanosine diphosphate L-fucose in porcine liver. J. Biol. Chem. 1968;243:1110–1115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

