Current Analytical Strategies for mRNA-Based Therapeutics
- PMID: 40286229
- PMCID: PMC11990077
- DOI: 10.3390/molecules30071629
Current Analytical Strategies for mRNA-Based Therapeutics
Abstract
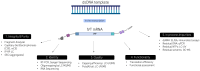
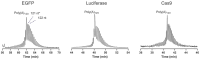
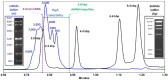
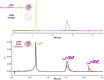
Recent advancements in mRNA technology, utilized in vaccines, immunotherapies, protein replacement therapies, and genome editing, have emerged as promising and increasingly viable treatments. The rapid, potent, and transient properties of mRNA-encoded proteins make them attractive tools for the effective treatment of a variety of conditions, ranging from infectious diseases to cancer and single-gene disorders. The capability for rapid and large-scale production of mRNA therapeutics fueled the global response to the COVID-19 pandemic. For effective clinical implementation, it is crucial to deeply characterize and control important mRNA attributes such as purity/integrity, identity, structural quality features, and functionality. This implies the use of powerful and advanced analytical techniques for quality control and characterization of mRNA. Improvements in analytical techniques such as electrophoresis, chromatography, mass spectrometry, sequencing, and functionality assessments have significantly enhanced the quality and detail of information available for product and process characterization, as well as for routine stability and release testing. Here, we review the latest advancements in analytical techniques for the characterization of mRNA-based therapeutics, typically employed by the biopharmaceutical industry for eventual market release.
Keywords: chromatography; electrophoresis; functionality; mass spectrometry; messenger RNA; quality attributes; sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
-
- Boros L.G., Kyriakopoulos A.M., Brogna C., Piscopo M., McCullough P.A., Seneff S. Long-lasting, biochemically modified mRNA, and its frameshifted recombinant spike proteins in human tissues and circulation after COVID-19 vaccination. Pharmacol. Res. Perspect. 2024;12:e1218. doi: 10.1002/prp2.1218. - DOI - PMC - PubMed
-
- Taguchi Y.-H. MicroRNA. Elsevier; Amsterdam, The Netherlands: 2022. RNA m6A Modification and microRNAs; pp. 169–180. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

