Identification of HIBCH and MGME1 as Mitochondrial Dynamics-Related Biomarkers in Alzheimer's Disease Via Integrated Bioinformatics Analysis
- PMID: 40286336
- PMCID: PMC12033025
- DOI: 10.1049/syb2.70018
Identification of HIBCH and MGME1 as Mitochondrial Dynamics-Related Biomarkers in Alzheimer's Disease Via Integrated Bioinformatics Analysis
Abstract
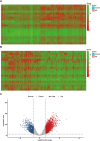
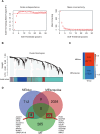
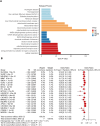
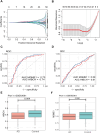
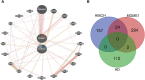
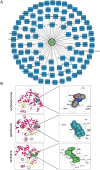
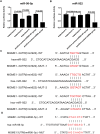
Mitochondrial dynamics (MD) play a crucial role in the genesis of Alzheimer's disease (AD); however, the molecular mechanisms underlying MD dysregulation in AD remain unclear. This study aimed to identify critical molecules of MD that contribute to AD progression using GEO data and bioinformatics approaches. The GSE63061 dataset comparing AD patients with healthy controls was analysed, WGCNA was employed to identify co-expression modules and differentially expressed genes (DEGs) and LASSO model was developed and verified using the DEGs to screen for potential biomarkers. A PPI network was built to predict upstream miRNAs, which were experimentally validated using luciferase reporter assays. A total of 3518 DEGs were identified (2209 upregulated, 1309 downregulated; |log2FC| > 1.5, adjusted p < 0.05). WGCNA revealed 160 MD-related genes. LASSO regression selected HIBCH and MGME1 as novel biomarkers with significant downregulation in AD (fold change > 2, p < 0.001). KEGG enrichment analysis highlighted pathways associated with neurodegeneration. Luciferase assays confirmed direct binding of miR-922 to the 3'UTR of MGME1. HIBCH and MGME1 are promising diagnostic biomarkers for AD with AUC values of 0.73 and 0.74. Mechanistically, miR-922 was experimentally validated to directly bind MGME1 3'UTR.
Keywords: bioinformatics; biomechanics; brain; data analysis; network analysis.
© 2025 The Author(s). IET Systems Biology published by John Wiley & Sons Ltd on behalf of The Institution of Engineering and Technology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

