Perturbing local steroidogenesis to improve breast cancer immunity
- PMID: 40287432
- PMCID: PMC12033260
- DOI: 10.1038/s41467-025-59356-3
Perturbing local steroidogenesis to improve breast cancer immunity
Abstract
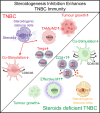
Breast cancer, particularly triple-negative breast cancer (TNBC), evades the body's immune defences, in part by cultivating an immunosuppressive tumour microenvironment. Here, we show that suppressing local steroidogenesis can augment anti-tumour immunity against TNBC. Through targeted metabolomics of steroids coupled with immunohistochemistry, we profiled the existence of immunosuppressive steroids in TNBC patient tumours and discerned the steroidogenic activity in immune-infiltrating regions. In mouse, genetic inhibition of immune cell steroidogenesis restricted TNBC tumour progression with a significant reduction in immunosuppressive components such as tumour associated macrophages. Steroidogenesis inhibition appears to bolster anti-tumour immune responses in dendritic and T cells by impeding glucocorticoid signalling. Undertaking metabolic modelling of the single-cell transcriptomics and targeted tumour-steroidomics, we pinpointed the predominant steroidogenic cells. Inhibiting steroidogenesis pharmacologically using a identified drug, posaconazole, curtailed tumour expansion in a humanised TNBC mouse model. This investigation paves the way for targeting steroidogenesis and its signalling pathways in breast cancer affected by immune-steroid maladaptation.
© 2025. Crown.
Conflict of interest statement
Competing interests: BM, JP and SKS declare following competing interests. An UK patent application submitted (Title: Cancer Treatments. Reference Number: P370182GB. Patent Application Number: 2502017.3) partly based on the findings in this manuscript where BM, JP and SKS are co-inventors. All other authors declare no competing interests.
Figures





References
-
- Giaquinto, A. N. et al. Breast cancer statistics, 2022. CA: A Cancer J. Clin.72, 524–541 (2022). - PubMed
-
- Carey, L., Winer, E., Viale, G., Cameron, D. & Gianni, L. Triple-negative breast cancer: disease entity or title of convenience? Nat. Rev. Clin. Oncol.7, 683–692 (2010). - PubMed
-
- Foulkes, W. D., Smith, I. E. & Reis-Filho, J. S. Triple-negative breast cancer. N. Engl. J. Med.363, 1938–1948 (2010). - PubMed
-
- Bianchini, G., De Angelis, C., Licata, L. & Gianni, L. Treatment landscape of triple-negative breast cancer—expanded options, evolving needs. Nat. Rev. Clin. Oncol.19, 91–113 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

