CLAVATA signalling shapes barley inflorescence by controlling activity and determinacy of shoot meristem and rachilla
- PMID: 40287461
- PMCID: PMC12033307
- DOI: 10.1038/s41467-025-59330-z
CLAVATA signalling shapes barley inflorescence by controlling activity and determinacy of shoot meristem and rachilla
Abstract
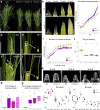
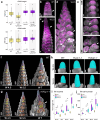
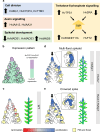
The large variety of inflorescence architectures evolved in grasses depends on shape, longevity and determinacy of meristems directing growth of the main and lateral axes. The CLAVATA pathway is known to regulate meristem size and inflorescence architecture in grasses. However, how individual meristem activities are determined and integrated to generate specific inflorescences is not yet understood. We found that activity of distinct meristems in the barley inflorescence is controlled by a signalling pathway comprising the receptor-like kinase Hordeum vulgare CLAVATA1 (HvCLV1) and the secreted CLAVATA3/EMBRYO-SURROUNDING REGION RELATED (CLE)-family peptide FON2-LIKE CLE PROTEIN1 (HvFCP1). HvFCP1 and HvCLV1 interact to promote spikelet formation, but restrict inflorescence meristem and rachilla proliferation. Hvfcp1 or Hvclv1 mutants generate additional rows of spikelets and supernumerary florets from extended rachilla activity. HvFCP1/HvCLV1 signalling coordinates meristem activity through regulation of trehalose-6-phosphate levels. Our discoveries outline a path to engineer inflorescence architecture via specific regulation of distinct meristem activities.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
INTERMEDIUM-M encodes an HvAP2L-H5 ortholog and is required for inflorescence indeterminacy and spikelet determinacy in barley.Proc Natl Acad Sci U S A. 2021 Feb 23;118(8):e2011779118. doi: 10.1073/pnas.2011779118. Proc Natl Acad Sci U S A. 2021. PMID: 33593903 Free PMC article.
-
Non-cell-autonomous signaling associated with barley ALOG1 specifies spikelet meristem determinacy.Curr Biol. 2024 Jun 3;34(11):2344-2358.e5. doi: 10.1016/j.cub.2024.04.083. Epub 2024 May 22. Curr Biol. 2024. PMID: 38781954
-
COMPOSITUM 1 contributes to the architectural simplification of barley inflorescence via meristem identity signals.Nat Commun. 2020 Oct 12;11(1):5138. doi: 10.1038/s41467-020-18890-y. Nat Commun. 2020. PMID: 33046693 Free PMC article.
-
Molecular Insights into Inflorescence Meristem Specification for Yield Potential in Cereal Crops.Int J Mol Sci. 2021 Mar 29;22(7):3508. doi: 10.3390/ijms22073508. Int J Mol Sci. 2021. PMID: 33805287 Free PMC article. Review.
-
Grass meristems II: inflorescence architecture, flower development and meristem fate.Plant Cell Physiol. 2013 Mar;54(3):313-24. doi: 10.1093/pcp/pct016. Epub 2013 Jan 31. Plant Cell Physiol. 2013. PMID: 23378448 Review.
Cited by
-
How meristems shape plant architecture in cereals-Cereal Stem Cell Systems (CSCS) Consortium.Plant Cell. 2025 Jul 1;37(7):koaf150. doi: 10.1093/plcell/koaf150. Plant Cell. 2025. PMID: 40578304 Free PMC article. Review.
References
-
- Zhang, D. & Yuan, Z. Molecular Control of Grass Inflorescence Development. Annu. Rev. Plant Biol.65, 553–578 (2014). - PubMed
-
- Koppolu, R. & Schnurbusch, T. Developmental pathways for shaping spike inflorescence architecture in barley and wheat. J. Integr. Plant Biol.61, 278–295 (2019). - PubMed
-
- Kyozuka, J., Tokunaga, H. & Yoshida, A. Control of grass inflorescence form by the fine-tuning of meristem phase change. Curr. Opin. Plant Biol.17, 110–115 (2014). - PubMed
-
- Bommert, P. & Whipple, C. Grass inflorescence architecture and meristem determinacy. Semin. Cell Dev. Biol.79, 37–47 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

