A feasibility of computational drug screening for Fuchs endothelial corneal dystrophy
- PMID: 40287480
- PMCID: PMC12033358
- DOI: 10.1038/s41598-025-95003-z
A feasibility of computational drug screening for Fuchs endothelial corneal dystrophy
Abstract
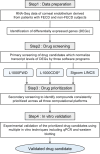
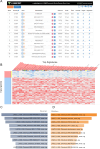
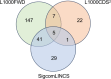
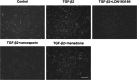
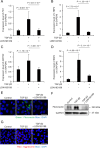
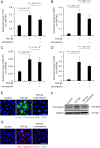
Fuchs endothelial corneal dystrophy (FECD) remains a leading cause of corneal blindness globally, with corneal transplantation being the primary treatment. FECD is characterized by the formation of guttae, extracellular matrix (ECM) deposits beneath the corneal endothelium, and progressive endothelial cell loss. These pathological changes cause visual deterioration through light scattering by guttae and corneal edema due to endothelial cell loss. However, limitations such as donor shortage and graft failure necessitate alternative therapeutic approaches. We employed computational drug screening using three platforms (L1000FWD, L1000CDS2, and SigCom LINCS) to identify compounds capable of normalizing FECD-associated differentially expressed genes (DEGs). Analysis of transcriptome data from FECD patients with TCF4trinucleotide repeat expansion identified 706 upregulated and 962 downregulated genes. The screening platforms identified 200, 35, and 76 compounds through L1000FWD, L1000CDS2, and SigCom LINCS, respectively, with five compounds commonly predicted across all platforms. Among these, LDN193189 and cercosporin were selected for further evaluation based on availability and lack of cytotoxicity. Both compounds significantly decreased the expression of ECM-related genes (FN1, MATN3, BGN, and LTBP2) in FECD cell models and suppressed TGF-β-induced fibronectin expression. Additionally, both compounds reduced aggresome formation to normal control levels, suggesting protection against endoplasmic reticulum stress-induced cell death. This study demonstrates the feasibility of computational drug screening for identifying therapeutic candidates for FECD, with LDN193189 and cercosporin showing promise in normalizing FECD-associated pathological changes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: Naoki Okumura and Noriko Koizumi are co-founders of ActualEyes Inc., which is currently developing a pharmaceutical therapy for treating Fuchs endothelial corneal dystrophy. Other authors do not have a competing financial interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

