Predictive models and WTAP targeting for idiopathic pulmonary fibrosis (IPF)
- PMID: 40287490
- PMCID: PMC12033295
- DOI: 10.1038/s41598-025-98490-2
Predictive models and WTAP targeting for idiopathic pulmonary fibrosis (IPF)
Abstract
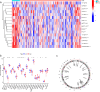
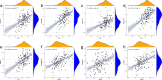
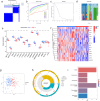
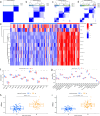
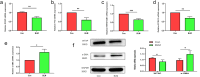
Emerging evidence suggests that N6-methyladenosine (m6A) modification significantly influences lung injury, lung cancer, and immune responses. The current study explores the potential involvement of m6A modification in the development of IPF. This research analyzed the GSE93606 dataset of 20 non-IPF and 154 IPF patients, identifying 26 m6A regulators and developing predictive models with RF and SVM, assessed via ROC curves. A nomogram was created with selected m6A factors, including molecular subtyping, PCA for m6A features, immune cell analysis, DEG identification, and functional enrichment. In vitro experiments on MRC-5 cells used RT-qPCR and Western blotting, and virtual drug screening targeted the WTAP protein through molecular docking. Analysis revealed 26 differential m6A regulators in IPF patients, with 16 significant; IGFBP2 and YTHDF2 were overexpressed, while others decreased. RF and SVM models identified predictive m6A regulators, and a nomogram was developed using five factors to predict IPF incidence. Distinct m6A patterns showed changes in RNA levels of specific genes in the BLM-induced group, and five compounds targeting WTAP were identified. This research explored m6A factors' impact on IPF diagnosis and prognosis, identifying WTAP as a potential biomarker.
Keywords: Idiopathic pulmonary fibrosis; Immune cell infiltration; Molecular docking; Prognostic biomarker; WTAP.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

