Characterization of transcriptomics during aging and genes required for lifespan in Drosophila intestine
- PMID: 40287511
- PMCID: PMC12033250
- DOI: 10.1038/s41598-025-98888-y
Characterization of transcriptomics during aging and genes required for lifespan in Drosophila intestine
Abstract
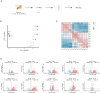
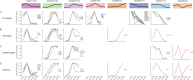
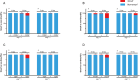
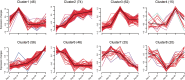
Aging is closely associated with imbalanced transcription. Regulated transcription in different organs is significantly different during aging, indicating that organ-specific transcriptomics is critical for understanding this process. Here we analyze the transcriptomics of the intestines of 3-, 15-, 30-, 40- and 50-days old female flies, which include young, middle-aged, and old flies. We find that the differential expression of protein-coding genes and lncRNAs is significant in aging, and fly age is characterized by well-separated gene expression trajectories. The highly clustered differentially expressed genes are connected to specific biological processes and signalling pathways. In particular, the Imd and Toll pathways are the top two immune signalling pathways that are highly regulated, and members with increased expression in the Imd pathway span all upstream activating events and include many ubiquitylation-associated factors and regulators of NF-κB factor Relish. Increased expression of Toll pathway members includes sensing mediators for all kinds of microorganisms and multiple proteases in the proteolytic processing cascade. Moreover, the expression of molecular markers of intestinal cells is greatly changed. Enterocyte markers are the most significantly influenced, and enteroendocrine markers AstA and NPF, as well as intestinal stem cell (ISC)/enteroblast (EB) markers Esg and Klu are expressed at low levels in young flies and much higher levels in aged flies. Furthermore, lncRNAs show similar expression trends and clustering patterns to those of protein-coding genes. Lastly, we find that ISC/EB-specific knock-down of 13 out of 19 genes that are highly differentially expressed reduces the lifespan of the fly. Together, the characterized transcriptomics and newly identified functional genes in aging will provide potential targets for preventing intestinal aging and associated disorders.
Keywords: Drosophila; Aging; Intestine; Lifespan; Transcriptomics; lncRNA.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

