Unveiling Berberine analogues as potential inhibitors of Escherichia coli FtsZ through machine learning molecular docking and molecular dynamics approach
- PMID: 40287515
- PMCID: PMC12033256
- DOI: 10.1038/s41598-025-98835-x
Unveiling Berberine analogues as potential inhibitors of Escherichia coli FtsZ through machine learning molecular docking and molecular dynamics approach
Abstract
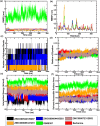
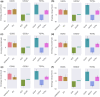
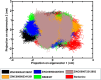
The bacterial cell division protein FtsZ, a crucial GTPase, plays a vital role in the formation of the contractile Z-ring, which is essential for bacterial cytokinesis. Consequently, inhibiting FtsZ could prevent the formation of proto-filaments and interfere with the cell division machinery. The remarkable conservation of FtsZ across diverse bacterial species makes it a promising drug target for combating drug resistance. In the present study, 1072 berberine analogues were screened for favorable pharmacokinetic properties. A total of 60 compounds that fulfilled the drug-likeliness criteria and were found to be non-toxic were selected for virtual screening against Escherichia coli FtsZ protein (PDB ID: 8GZY). Molecular docking revealed a strong binding affinity of ZINC000524729297 (- 8.73 kcal/mol) and ZINC000604405393 (and - 8.55 kcal/mol) with FtsZ by strong intermolecular hydrogen bonds and hydrophobic interactions. Subsequently, the docking profiles were validated through a 500 ns MD simulation and MMPBSA analysis of the FtsZ-ligand complexes. The analysis revealed the FtsZ- ZINC524729297 and FtsZ-ZINC000604405393 complexes had the lowest root-mean-square deviation with lowest binding energy and enhanced conformational stability in a dynamic environment. These findings suggest that ZINC524729297 and ZINC000604405393 are the potent lead compound that targets FtsZ and requires further experimental validation.
Keywords: Escherichia coli; Antimicrobial resistance; FtsZ; Machine learning; Molecular docking; Molecular dynamics simulation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




References
-
- Owrangi, B. et al. Invasion and translocation of uropathogenic Escherichia coli isolated from Urosepsis and patients with community-acquired urinary tract infection. Eur. J. Clin. Microbiol. Infect. Dis.37, 833–839 (2018). - PubMed
-
- Miajlovic, H., Mac Aogáin, M., Collins, C. J., Rogers, T. R. & Smith, S. G. J. Characterization of Escherichia coli bloodstream isolates associated with mortality. J. Med. Microbiol.65, 71–79 (2016). - PubMed
-
- Köhler, C. D. & Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol.301, 642–647 (2011). - PubMed
-
- Usein, C. R., Papagheorghe, R., Oprea, M., Condei, M. & Strãuţ, M. Molecular characterization of bacteremic Escherichia coli isolates in Romania. Folia Microbiol. (Praha). 61, 221–226 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

