Conservation genomics of a threatened subtropical Rhododendron species highlights the distinct conservation actions required in marginal and admixed populations
- PMID: 40287966
- PMCID: PMC12034323
- DOI: 10.1111/tpj.70175
Conservation genomics of a threatened subtropical Rhododendron species highlights the distinct conservation actions required in marginal and admixed populations
Abstract
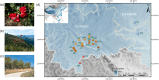
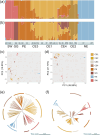
With the impact of climate change and anthropogenic activities, the underlying threats facing populations with different evolutionary histories and distributions, and the associated conservation strategies necessary to ensure their survival, may vary within a species. This is particularly true for marginal populations and/or those showing admixture. Here, we re-sequence genomes of 102 individuals from 21 locations for Rhododendron vialii, a threatened species distributed in the subtropical forests of southwestern China that has suffered from habitat fragmentation due to deforestation. Population structure results revealed that R. vialii can be divided into five genetic lineages using neutral single-nucleotide polymorphisms (SNPs), whereas selected SNPs divide the species into six lineages. This is due to the Guigu (GG) population, which is identified as admixed using neutral SNPs, but is assigned to a distinct genetic cluster using non-neutral loci. R. vialii has experienced multiple genetic bottlenecks, and different demographic histories have been suggested among populations. Ecological niche modeling combined with genomic offset analysis suggests that the marginal population (Northeast, NE) harboring the highest genetic diversity is likely to have the highest risk of maladaptation in the future. The marginal population therefore needs urgent ex situ conservation in areas where the influence of future climate change is predicted to be well buffered. Alternatively, the GG population may have the potential for local adaptation, and will need in situ conservation. The Puer population, which carries the heaviest genetic load, needs genetic rescue. Our findings highlight how population genomics, genomic offset analysis, and ecological niche modeling can be integrated to inform targeted conservation.
Keywords: Rhododendron; climate change; conservation management; demographic history; genomic offset; local adaptation.
© 2025 The Author(s). The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Aguirre‐Liguori, J.A. , Ramírez‐Barahona, S. & Gaut, B.S. (2021) The evolutionary genomics of species' responses to climate change. Nature Ecology & Evolution, 5, 1350–1360. - PubMed
-
- Allendorf, F.W. (2017) Genetics and the conservation of natural populations: allozymes to genomes. Molecular Ecology, 26, 420–430. - PubMed
MeSH terms
Grants and funding
- 2024YFF1307400/National Key Research and Development Program of China
- 2019QZKK0502/Second Tibetan Plateau Scientific Expedition and Research Program
- 202403AC100028/Key R&D Program of Yunnan Province
- 2022SJ07X-03/Conservation Grant for PSESP in Yunnan Province
- 202404BI090014/Yunnan Provincial Science and Technology Mission
LinkOut - more resources
Full Text Sources

