High-resolution analysis of the treated coeliac disease microbiome reveals strain-level variation
- PMID: 40289251
- PMCID: PMC12036492
- DOI: 10.1080/19490976.2025.2489071
High-resolution analysis of the treated coeliac disease microbiome reveals strain-level variation
Abstract
Background: Coeliac disease (CeD) is an immune-mediated disorder primarily affecting the small intestine, characterized by an inflammatory immune reaction to dietary gluten. CeD onset results from a multifaceted interplay of genetic and environmental factors. While recent data show that alterations in gut microbiome composition could play an important role, many current studies are constrained by small sample sizes and limited resolution.
Methods: To address these limitations, we analyzed fecal gut microbiota from two Dutch cohorts, CeDNN (128 treated CeD patients (tCeD), 106 controls) and the Lifelines Dutch Microbiome Project (24 self-reported tCeD, 654 controls), using shotgun metagenomic sequencing. Self-reported IBS (570 cases, 1710 controls) and IBD (93 cases, 465 controls) were used as comparative conditions of the gastrointestinal tract. Interindividual variation within the case and control groups was calculated at whole microbiome and strain level. Finally, species-specific gene repertoires were analyzed in tCeD patients and controls.
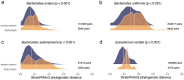
Results: Within-individual microbiome diversity was decreased in patients with self-reported IBS and IBD but not in tCeD patients. Each condition displayed a unique microbial pattern and, in addition to confirming previously reported microbiome associations, we identify an increase in the levels of Clostridium sp. CAG:253, Roseburia hominis, and Eggerthella lenta, amongst others. We further show that the observed changes can partially be explained by gluten-free diet adherence. We also observe increased interindividual variation of gut microbiome composition among tCeD patients and a higher bacterial mutation frequency in tCeD that contributes to higher interindividual variation at strain level. In addition, the immotile European subspecies of Eubacterium rectale, which has a distinct carbohydrate metabolism potential, was nearly absent in tCeD patients.
Conclusion: Our study sheds light on the complex interplay between the gut microbiome and CeD, revealing increased interindividual variation and strain-level variation in tCeD patients. These findings expand our understanding of the microbiome's role in intestinal health and disease.
Keywords: Coeliac disease; gluten; gluten-free diet; gut microbiome; metagenomic sequencing.
Conflict of interest statement
A.Z. received a speaker fee from Nestle. R.K.W. acted as a consultant for Takeda and received unrestricted research grants from Takeda and Johnson and Johnson pharmaceuticals and speaker fees from AbbVie, MSD, Olympus and AstraZeneca. The funders had no role in study design, data analysis, data interpretation, writing of the manuscript, and the decision to publish. Other authors have no potential conflicts of interest to report.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
