Gut microbes-spinal connection is required for itch sensation
- PMID: 40289281
- PMCID: PMC12036491
- DOI: 10.1080/19490976.2025.2495859
Gut microbes-spinal connection is required for itch sensation
Abstract
The gut microbiota has been linked to a number of neurological disorders. However, it is unclear whether the gut microbiota is involved in the genesis of chronic itch, a refractory condition that afflicts patients both physically and mentally. Here, we report that depletion of gut microbiota enhances tolerance to itch in mice orally administered with antibiotics (ABX) and mice free of germ. Of note, oral gavage with Bacteroides fragilis (B. fragilis), a prominent species of the genus Bacteroides with most differential change, corrected the ABX-induced itch dysfunction through its driven metabolite acetyl-l-carnitine (ALC). Mechanistically, gut microbiota or B. fragilis depletion caused a decrease in RNA N6-methyladenosine (m6A) demethylase FTO expression in the dorsal horn and a consequent increase in RNA m6A sites in Mas-related G protein-coupled receptor F (MrgprF) mRNA, leading to decreased MRGPRF protein. The downregulation of FTO was triggered by inactivation of ETS proto-oncogene 1 (ETS1), a transcription factor that binds to the Fto promoter. These findings support a gut microbe - spinal connection in modulation of itch sensation in RNA m6A epigenetic-dependent manner and highlight a critical role of ALC in linking the altered B. fragilis and itch dysfunction.
Keywords: B. fragilis; FTO; Gut microbe; RNA m6A; acetyl-l-carnitine; itch.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
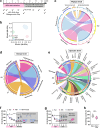
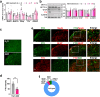



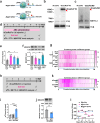



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
