An approach to building foundation models for brain image analysis
- PMID: 40290346
- PMCID: PMC12033034
- DOI: 10.1007/978-3-031-72390-2_40
An approach to building foundation models for brain image analysis
Abstract
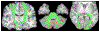
Existing machine learning methods for brain image analysis are mostly based on supervised training. They require large labeled datasets, which can be costly or impossible to obtain. Moreover, the trained models are useful only for the narrow task defined by the labels. In this work, we developed a new method, based on the concept of foundation models, to overcome these limitations. Our model is an attention-based neural network that is trained using a novel self-supervised approach. Specifically, the model is trained to generate brain images in a patch-wise manner, thereby learning the brain structure. To facilitate learning of image details, we propose a new method that encodes high-frequency information using convolutional kernels with random weights. We trained our model on a pool of 10 public datasets. We then applied the model on five independent datasets to perform segmentation, lesion detection, denoising, and brain age estimation. Results showed that the foundation model achieved competitive or better results on all tasks, while significantly reducing the required amount of labeled training data. Our method enables leveraging large unlabeled neuroimaging datasets to effectively address diverse brain image analysis tasks and reduce the time and cost requirements of acquiring labels.
Keywords: brain; deep learning; foundation models; neuroimaging.
Conflict of interest statement
Disclosure of Interests. The author has no competing interests to declare that are relevant to the content of this article.
Figures
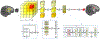
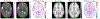


Similar articles
-
Local contrastive loss with pseudo-label based self-training for semi-supervised medical image segmentation.Med Image Anal. 2023 Jul;87:102792. doi: 10.1016/j.media.2023.102792. Epub 2023 Mar 11. Med Image Anal. 2023. PMID: 37054649
-
Semi-supervised abdominal multi-organ segmentation by object-redrawing.Med Phys. 2024 Nov;51(11):8334-8347. doi: 10.1002/mp.17364. Epub 2024 Aug 21. Med Phys. 2024. PMID: 39167059
-
Ultrasound carotid plaque segmentation via image reconstruction-based self-supervised learning with limited training labels.Math Biosci Eng. 2023 Jan;20(2):1617-1636. doi: 10.3934/mbe.2023074. Epub 2022 Nov 3. Math Biosci Eng. 2023. PMID: 36899501
-
Learning low-dose CT degradation from unpaired data with flow-based model.Med Phys. 2022 Dec;49(12):7516-7530. doi: 10.1002/mp.15886. Epub 2022 Aug 8. Med Phys. 2022. PMID: 35880375
-
Semi Supervised Learning with Deep Embedded Clustering for Image Classification and Segmentation.IEEE Access. 2019;7:11093-11104. doi: 10.1109/ACCESS.2019.2891970. Epub 2019 Jan 9. IEEE Access. 2019. PMID: 31588387 Free PMC article.
References
-
- Assran M, et al.: Masked siamese networks for label-efficient learning. In: European Conference on Computer Vision. pp. 456–473. Springer; (2022)
-
- Basri R, et al.: Frequency bias in neural networks for input of non-uniform density. In: International Conference on Machine Learning. pp. 685–694. PMLR; (2020)
-
- Baur C, et al.: Deep autoencoding models for unsupervised anomaly segmentation in brain mr images. In: 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018. pp. 161–169. Springer; (2019)
-
- Baur C, et al.: Autoencoders for unsupervised anomaly segmentation in brain mr images: a comparative study. Medical Image Analysis 69, 101952 (2021) - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
