Standardization of a High-Quality Methodological Framework for Long-Term Genetic Monitoring of the French Wolf Population
- PMID: 40290392
- PMCID: PMC12022234
- DOI: 10.1002/ece3.71345
Standardization of a High-Quality Methodological Framework for Long-Term Genetic Monitoring of the French Wolf Population
Abstract
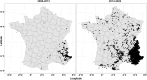
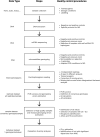
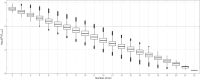
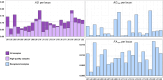
Since the gray wolf was eradicated from large parts of Europe, this species has been recolonizing much of its former distribution, particularly since the past 30 years. Wolves benefit from European legal protection through the Habitats Directive and the Bern Convention, and reporting on the evolution of their populations in each country of Europe is mandatory. To monitor French wolf populations over the long term, a standardized high-quality methodological framework has been developed to analyze data from noninvasively collected samples and assess population diversity. We delineated each step and implemented a laboratory control procedure to analyze 8733 samples harvested within the French distribution range of the species between 2006 and 2022, and provided key quality and diversity indicators. Of these samples, 82.8% were successfully amplified and sequenced for the mitochondrial control region. Subsequently, the wolf samples were genotyped at 22 microsatellite autosomal loci and a sex locus displayed over two independent multiplexes using the multitube approach. The average success rate of polymerase chain reaction per locus was 64.2% across all replicates. The residual genotyping error rates were low compared to those in other studies using non-invasively collected samples, with mean residual allelic dropout rates of 5.8% per locus and mean residual false allele rates of 1.0% per locus. The high-quality dataset identified 1735 individuals in total over the last 15 years, of which 99.9% exhibited a single Italo-Alpine mitotype. Genetic diversity was relatively low, with mean observed heterozygosity of 0.482 and mean expected heterozygosity of 0.519. This supports the natural colonization of the French Alps by a few individuals originating from the remaining Italian populations, which started approximately 30 years ago. By generating high-quality standards and quality control processes, this protocol enhances the cost-efficiency ratio of monitoring French wolf populations and holds high value for managers tasked with the management and conservation of wolf populations in the long term.
Keywords: Canis lupus; France; gray wolf; long‐term monitoring; microsatellite; mitochondrial control region; noninvasive samples.
© 2025 The Author(s). Ecology and Evolution published by British Ecological Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
From the Apennines to the Alps: colonization genetics of the naturally expanding Italian wolf (Canis lupus) population.Mol Ecol. 2007 Apr;16(8):1661-71. doi: 10.1111/j.1365-294X.2007.03262.x. Mol Ecol. 2007. PMID: 17402981
-
Noninvasive molecular tracking of colonizing wolf (Canis lupus) packs in the western Italian Alps.Mol Ecol. 2002 May;11(5):857-68. doi: 10.1046/j.1365-294x.2002.01489.x. Mol Ecol. 2002. PMID: 11975702
-
Wolves Recolonizing Islands: Genetic Consequences and Implications for Conservation and Management.PLoS One. 2016 Jul 6;11(7):e0158911. doi: 10.1371/journal.pone.0158911. eCollection 2016. PLoS One. 2016. PMID: 27384049 Free PMC article.
-
Evidence of genetic distinction and long-term population decline in wolves (Canis lupus) in the Italian Apennines.Mol Ecol. 2004 Mar;13(3):523-36. doi: 10.1046/j.1365-294x.2004.02077.x. Mol Ecol. 2004. PMID: 14871358
-
Helminth parasites of wolves (Canis lupus): a species list and an analysis of published prevalence studies in Nearctic and Palaearctic populations.J Helminthol. 2005 Jun;79(2):95-103. doi: 10.1079/joh2005282. J Helminthol. 2005. PMID: 15946392 Review.
References
-
- Beja‐Pereira, A. , Oliveira R., Alves P. C., Schwartz M. K., and Luikart G.. 2009. “Advancing Ecological Understandings Through Technological Transformations in Noninvasive Genetics.” Molecular Ecology Resources 9: 1279–1301. - PubMed
-
- Bonin, A. , Bellemain E., Bronken Eidesen P., Pompanon F., Brochmann C., and Taberlet P.. 2004. “How to Track and Assess Genotyping Errors in Population Genetics Studies.” Molecular Ecology 13: 3261–3273. - PubMed
-
- Broquet, T. , and Petit E.. 2004. “Quantifying Genotyping Errors in Noninvasive Population Genetics.” Molecular Ecology 13: 3601–3608. - PubMed
-
- Campbell, N. R. , Harmon S. A., and Narum S. R.. 2015. “Genotyping‐In‐Thousands by Sequencing (GT‐Seq): A Cost Effective SNP Genotyping Method Based on Custom Amplicon Sequencing.” Molecular Ecology Resources 15: 855–867. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

