Ultrasensitive RNase H activity detection using the transcription-based hybrid probe and CRISPR/cas12a signal amplifier
- PMID: 40290436
- PMCID: PMC12021801
- DOI: 10.3389/fphar.2025.1589150
Ultrasensitive RNase H activity detection using the transcription-based hybrid probe and CRISPR/cas12a signal amplifier
Abstract
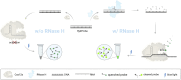
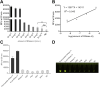
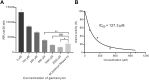
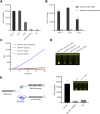
Ribonuclease H (RNase H), a critical functional protein in replication and genome stability, is emerging as a crucial therapeutic target for various diseases, including immune disorders. We present a transcription-based hybrid probe, referred to as Hybprobe, and a CRISPR/Cas12a signal amplifier for the rapid, sensitive, and low-cost detection of RNase H activity. In this method, the RNA strand of the Hybprobe is specifically cleaved by RNase H, releasing a single-stranded DNA activator that facilitates recognition and cleavage by the Cas12a/crRNA complex, triggering signal amplification via Cas12a's trans-cleavage activity. The proposed method demonstrates ultra-high sensitivity, capable of detecting RNase H as low as 9.02 × 10-10 U/μL, making it approximately 1,000 times more sensitive than several previously reported methods. Furthermore, we demonstrated the application of this method for RNase H inhibitor evaluation and its practical use across various biological samples, including cell extracts and HIV reverse transcriptase. In summary, the results suggest that this method is a promising tool for the highly sensitive detection of RNase H and the diagnosis of diseases associated with RNase H.
Keywords: CRISPR/Cas12a; Rnase H; activity detection; hybprobe; inhibitor screening.
Copyright © 2025 Ding, Wei, Yang, Shi, Ren, Li and Tang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
A novel fluorescence amplification strategy combining cascade primer exchange reaction with CRISPR/Cas12a system for ultrasensitive detection of RNase H activity.Biosens Bioelectron. 2022 Jun 15;206:114135. doi: 10.1016/j.bios.2022.114135. Epub 2022 Mar 4. Biosens Bioelectron. 2022. PMID: 35278851
-
An amplification-free CRISPR/Cas12a-based fluorescence assay for ultrasensitive detection of nuclease activity.Talanta. 2023 May 15;257:124329. doi: 10.1016/j.talanta.2023.124329. Epub 2023 Feb 8. Talanta. 2023. PMID: 36801553
-
Controllable crRNA Self-Transcription Aided Dual-Amplified CRISPR-Cas12a Strategy for Highly Sensitive Biosensing of FEN1 Activity.ACS Synth Biol. 2022 Nov 18;11(11):3847-3854. doi: 10.1021/acssynbio.2c00420. Epub 2022 Oct 14. ACS Synth Biol. 2022. PMID: 36240131
-
Exploring the trans-cleavage activity of CRISPR-Cas12a for the development of a Mxene based electrochemiluminescence biosensor for the detection of Siglec-5.Biosens Bioelectron. 2021 Apr 15;178:113019. doi: 10.1016/j.bios.2021.113019. Epub 2021 Jan 26. Biosens Bioelectron. 2021. PMID: 33517231
-
Hybridization chain reaction circuit controller: CRISPR/Cas12a conversion amplifier for miRNA-21 sensitive detection.Talanta. 2024 Jan 1;266(Pt 2):125130. doi: 10.1016/j.talanta.2023.125130. Epub 2023 Aug 26. Talanta. 2024. PMID: 37657377
References
LinkOut - more resources
Full Text Sources

