A Pyroptosis-Related LncRNA Signature for Predicting Prognosis, Immune Features and Drug Sensitivity in Ovarian Cancer
- PMID: 40291608
- PMCID: PMC12034292
- DOI: 10.2147/OTT.S491130
A Pyroptosis-Related LncRNA Signature for Predicting Prognosis, Immune Features and Drug Sensitivity in Ovarian Cancer
Abstract
Background: Multiple studies have suggested that lncRNAs and pyroptosis play important roles in ovarian cancer (OC). However, the function of pyroptosis-related lncRNAs (PRLs) in OC is not fully understood.
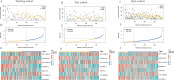
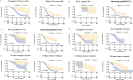
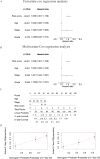
Methods: Clinical information and RNA-seq data of OC patients (n = 379) were collected from TCGA database. Pearson correlation analysis and univariate Cox analysis were performed to identify prognostic PRLs, respectively. LASSO-COX regression was utilized to construct a prognostic PRLs signature. Kaplan-Meier (K-M) curve analyses and receiver operating characteristics (ROC) were used to evaluate the prognostic prediction of the signature. The association between risk score and tumor microenvironment infiltration, immunotherapy response and chemotherapy sensitivity were also analyzed. In addition, the function of TYMSOS on OC and pyroptosis was experimentally confirmed in cell lines.
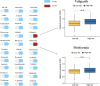
Results: Firstly, 32 prognostic PRLs were identified, and a novel prognostic PRLs signature was constructed and validated. Surprisingly, the prognostic PRLs signature could solidly predict the clinical outcome of patients with OC and patients with high-risk score shown a short overall survival. GSEA results suggested that the RPLs were mainly enriched in the inflammatory response pathway, p53 pathway, TGF-β signaling and TNFα signaling. Besides, our results demonstrated that the risk score was significantly associated with patients with immune infiltration, immunotherapy response and the sensitivity of veliparib and metformin. Furthermore, the oncogene effect of TYMSOS on OC by inhibiting pyroptosis was verified by experiments.
Conclusion: This study found that the prognostic PRLs signature may serve as an efficient biomarker in predicting the prognosis, tumor microenvironment infiltration, and sensitivity of chemotherapeutic agents. TYMSOS is a potential biomarker in OC, and it might promote tumor progression by inhibiting pyroptosis.
Keywords: immune microenvironment; ovarian cancer; prognosis; pyroptosis; signature.
© 2025 Liu et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
-
- Koren E, Fuchs Y. Modes of regulated cell death in cancer. Cancer Discov. 2021;11(2):245–265. doi: 10.1158/2159-8290.CD-20-0789 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

