Changes in the community composition and function of the rhizosphere microbiome in tobacco plants with Fusarium root rot
- PMID: 40291803
- PMCID: PMC12023262
- DOI: 10.3389/fmicb.2025.1512694
Changes in the community composition and function of the rhizosphere microbiome in tobacco plants with Fusarium root rot
Abstract
Introduction: Tobacco root rot caused by Fusarium spp. is a soil-borne vascular disease that severely affects tobacco production worldwide. To date, the community composition and functional shifts of the rhizosphere microbiome in tobacco plants infected with Fusarium root rot remain poorly understood.
Methods: In this study, we analyzed the differences in the compositions and functions of the bacterial and fungal communities in the rhizosphere and root endosphere of healthy tobacco plants and tobacco with Fusarium root rot using amplicon sequencing and metagenomic sequencing.
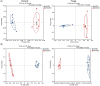
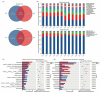
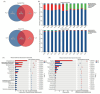
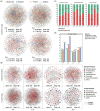
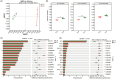
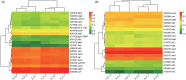
Results and discussion: Our results showed that Fusarium root rot disrupted the stability of bacteria-fungi interkingdom networks and reduced the network complexity. Compared to healthy tobacco plants, the Chao1 index of bacterial communities in the rhizosphere soil of diseased plants increased by 4.09% (P < 0.05), while the Shannon and Chao1 indices of fungal communities decreased by 13.87 and 8.17%, respectively (P < 0.05). In the root tissues of diseased plants, the Shannon index of bacterial and fungal communities decreased by 17.71-27.05% (P < 0.05). Additionally, we observed that the rhizosphere microbial community of diseased tobacco plants shifted toward a pathological combination, with a significant increase in the relative abundance of harmful microbes such as Alternaria, Fusarium, and Filobasidium (89.46-921.29%) and a notable decrease in the relative abundance of beneficial microbes such as Lysobacter, Streptomyces, Mortierella, and Penicillium (48.48-81.56%). Metagenomic analysis further revealed that the tobacco rhizosphere microbial communities of diseased plants played a significant role in basic biological metabolism, energy production and conversion, signal transduction, and N metabolism, but their functions involved in C metabolism were significantly weakened. Our findings provide new insights into the changes in and interactions within the rhizosphere and root endosphere microbiomes of tobacco plants under the stress of Fusarium soil-borne fungal pathogens, while laying the foundation for the exploration, development, and utilization of beneficial microbial resources in healthy tobacco plants in the future.
Keywords: Fusarium spp.; microbial diversity; microbial interactions; rhizosphere; soil-borne fungal disease.
Copyright © 2025 Yang, Cai, Bai, Han, Zeng, Liu, Liu, Liu, Ma and Yu.
Conflict of interest statement
YC, TB, RZ, DL, TL, RL, and CM were employed by Qujing Branch of Yunnan Provincial Tobacco Company. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Ahmed W., Dai Z., Liu Q., Munir S., Yang J., Karunarathna S. C., et al. (2022). Microbial cross-talk: dissecting the core microbiota associated with flue-cured tobacco (Nicotiana tabacum) plants under healthy and diseased state. Front. Microbiol. 13:845310. 10.3389/fmicb.2022.845310 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

