Characterization of the novel cross-genus phage vB_SmaS_QH3 and evaluation of its antibacterial efficacy against Stenotrophomonas maltophilia
- PMID: 40291807
- PMCID: PMC12023781
- DOI: 10.3389/fmicb.2025.1570665
Characterization of the novel cross-genus phage vB_SmaS_QH3 and evaluation of its antibacterial efficacy against Stenotrophomonas maltophilia
Abstract
Background: Bacteriophages, which are natural bacterial predators, demonstrate potential as safe and effective biological control agents against drug-resistant infections. This study aims to characterize the biological properties of the novel lytic phage vB_SmaS_QH3 and comprehensively evaluate its efficacy in preventing and controlling clinically multidrug resistance Stenotrophomonas maltophilia infections using both in vivo and in vitro models.
Methods: The phage was isolated from hospital sewage using the multidrug resistant S. maltophilia no. 3738 as the host. Transmission electron microscopy (TEM) was used to observe phage morphology, and the host range was determined via spot assays. Proliferation kinetics, including multiplicity of infection (MOI), adsorption rate, and one-step growth curves, were analyzed. Stability was assessed under various physicochemical conditions. Based on Illumina whole-genome sequencing data, bioinformatics tools were employed for gene annotation, functional prediction, and phylogenetic analysis. Antimicrobial activity was assessed using in vitro and in vivo models.
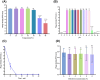
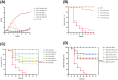
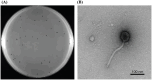
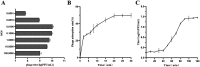
Results: A lytic phage vB_SmaS_QH3 was isolated from hospital sewage. TEM revealed that it belongs to the class Caudoviricetes, featuring an icosahedral head (62 ± 3 nm) and a non-contractile long tail (121 ± 5 nm). Although the phage has a narrow host range, it exhibits cross-genus infectivity, lysing S. maltophilia (11/81) and Pseudomonas aeruginosa (3/24). The optimal MOI for phage vB_SmaS_QH3 is 0.01, with an adsorption rate of 49.16% within 20 min, a latent period of 40 min, a lytic period of 50 min, and a burst size of 41.67 plaque-forming units/cell. The phage remained stable at 4-60°C, at pH 3-11, and in chloroform, but it was completely inactivated following 20-min exposure to UV irradiation. Genomic analysis showed a linear double-stranded DNA genome of 43,085 bp with a GC content of 54.2%, containing 54 predicted ORFs, and no virulence or antibiotic resistance genes were detected. In vitro, vB_SmaS_QH3 effectively inhibited bacterial growth within 9 h. In vivo, it significantly improved the survival rate of Galleria mellonella larvae infected with S. maltophilia, regardless of the treatment timing.
Conclusion: vB_SmaS_QH3 is a narrow host range lytic phage with a safe genome and excellent stability. It exhibits significant antibacterial activity both in vitro and in vivo, making it a promising candidate for therapeutic applications.
Keywords: Galleria mellonella; Stenotrophomonas maltophilia; genomic analysis; phage; phage therapy.
Copyright © 2025 Cheng, Li, Liu, Li, Zhou, Luo, Mu, Sun, Ma and A.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Alcock B. P., Huynh W., Chalil R., Smith K. W., Raphenya A. R., Wlodarski M. A., et al. (2022). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucl. Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

