Muscle mTOR controls iron homeostasis and ferritinophagy via NRF2, HIFs and AKT/PKB signaling pathways
- PMID: 40293459
- PMCID: PMC12037468
- DOI: 10.1007/s00018-025-05695-9
Muscle mTOR controls iron homeostasis and ferritinophagy via NRF2, HIFs and AKT/PKB signaling pathways
Abstract
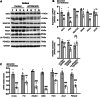
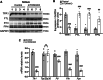
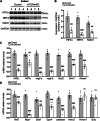
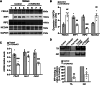
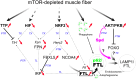
Balanced mTOR activity and iron levels are crucial for muscle integrity, with evidence suggesting mTOR regulates cellular iron homeostasis. In this study, we investigated iron metabolism in muscle-specific mTOR knockout mice (mTORmKO) and its relation to their myopathy. The mTORmKO mice exhibited distinct iron content patterns across muscle types and ages. Slow-twitch soleus muscles initially showed reduced iron levels in young mice, which increased with the dystrophy progression but remained within control ranges. In contrast, the less affected fast-twitch muscles maintained near-normal iron levels from a young age. Interestingly, both mTORmKO muscle types exhibited iron metabolism markers indicative of iron excess, including decreased transferrin receptor 1 (TFR1) and increased levels of ferritin (FTL) and ferroportin (FPN) proteins. Paradoxically, these changes were accompanied by downregulated Ftl and Fpn mRNA levels, indicating post-transcriptional regulation. This discordant regulation resulted from disruption of key iron metabolism pathways, including NRF2/NFE2L2, HIFs, and AKT/PKB signaling. Mechanistically, mTOR deficiency impaired transcriptional regulation of iron-related genes mediated by NRF2 and HIFs. Furthermore, it triggered ferritin accumulation through two NRF2 mechanisms: (1) derepression of ferritin translation via suppression of the FBXL5-IRP axis, and (2) autophagosomal sequestration driven by NCOA4-dependent ferritin targeting to autophagosomes, coupled with age-related impairments of autophagy linked to chronic AKT/PKB activation. Three-week spermidine supplementation in older mTORmKO mice was associated with normalized AKT/PKB-FOXO signaling, increased endolysosomal FTL and reduced total FTL levels in the dystrophic soleus muscle. These findings underscore mTOR's crucial role in skeletal muscle iron metabolism and suggest spermidine as a potential strategy to address impaired ferritinophagy due to autophagy blockade in dystrophic muscle.
Keywords: Autophagy; Dystrophy; Glycogen; Iron-sulfur cluster; Myoglobin; Oxidative stress.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: Experimental protocols involving animals were performed in accordance with national and European legislation on animal experimentation and were approved by the local animal ethics committee (Apafis#18195; Ceccapp-ENS_2013_013; Ceccapp-ENS_2011_030). Consent to participate: Not applicable. Consent for publication: Not applicable. Conflict of interest: The authors declare no conflict of interest.
Figures


Similar articles
-
Lack of muscle mTOR kinase activity causes early onset myopathy and compromises whole-body homeostasis.J Cachexia Sarcopenia Muscle. 2019 Feb;10(1):35-53. doi: 10.1002/jcsm.12336. Epub 2018 Nov 21. J Cachexia Sarcopenia Muscle. 2019. PMID: 30461220 Free PMC article.
-
NCOA4-Mediated Ferritinophagy Is a Pancreatic Cancer Dependency via Maintenance of Iron Bioavailability for Iron-Sulfur Cluster Proteins.Cancer Discov. 2022 Sep 2;12(9):2180-2197. doi: 10.1158/2159-8290.CD-22-0043. Cancer Discov. 2022. PMID: 35771492 Free PMC article.
-
Plasticity changes in iron homeostasis in hibernating Daurian ground squirrels (Spermophilus dauricus) may counteract chronically inactive skeletal muscle atrophy.J Comp Physiol B. 2024 Apr;194(2):191-202. doi: 10.1007/s00360-024-01543-7. Epub 2024 Mar 24. J Comp Physiol B. 2024. PMID: 38522042
-
The Role of NCOA4-Mediated Ferritinophagy in Ferroptosis.Adv Exp Med Biol. 2021;1301:41-57. doi: 10.1007/978-3-030-62026-4_4. Adv Exp Med Biol. 2021. PMID: 34370287 Review.
-
Molecular regulation of cellular iron balance.IUBMB Life. 2017 Jun;69(6):389-398. doi: 10.1002/iub.1628. Epub 2017 May 7. IUBMB Life. 2017. PMID: 28480557 Review.
References
-
- Xu J, Hwang JC, Lees HA, Wohlgemuth SE, Knutson MD, Judge AR et al (2012) Long-term perturbation of muscle iron homeostasis following hindlimb suspension in old rats is associated with high levels of oxidative stress and impaired recovery from atrophy. Exp Gerontol 47:100–108. 10.1016/j.exger.2011.10.011 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

