Meiotic divisions and round spermatid formation do not require centriole duplication in mice
- PMID: 40294089
- PMCID: PMC12064039
- DOI: 10.1371/journal.pgen.1011698
Meiotic divisions and round spermatid formation do not require centriole duplication in mice
Abstract
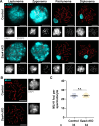
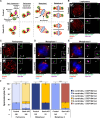
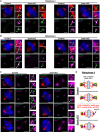
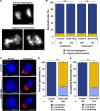
Centrosomes, composed of centrioles and pericentriolar matrix proteins, are traditionally viewed as essential microtubule-organizing centers (MTOCs) that facilitate bipolar spindle formation and chromosome segregation during spermatogenesis. In this study, we investigated the role of centrioles in male germ cell development by using a murine conditional knockout (cKO) of Sas4, a critical component of centriole biogenesis. We found that while centriole duplication was impaired in Sas4 cKO spermatocytes, these cells were still capable of progressing through meiosis I and II. Chromosome segregation was able to proceed through the formation of a non-centrosomal MTOC, indicating that centrioles are not required for meiotic divisions. However, spermatids that inherited fewer than two centrioles exhibited severe defects in spermiogenesis, including improper manchette formation, constricted perinuclear rings, disrupted acrosome morphology, and failure to form flagella. Consequently, Sas4 cKO males were infertile due to the absence of functional spermatozoa. Our findings demonstrate that while centrioles are dispensable for meiosis in male germ cells, they are essential for spermiogenesis and sperm maturation. This work provides key insights into the role of centrosomes in male fertility and may have implications for understanding certain conditions of male infertility associated with centriole defects.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: PWJ is on the scientific advisory board of Gameto, Inc. All other authors have no competing interests to disclose.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

