Epigenetic silencing of DNA sensing pathway by FOXM1 blocks stress ligand-dependent antitumor immunity and immune memory
- PMID: 40295473
- PMCID: PMC12037779
- DOI: 10.1038/s41467-025-59186-3
Epigenetic silencing of DNA sensing pathway by FOXM1 blocks stress ligand-dependent antitumor immunity and immune memory
Abstract
The interplay between tumor cells and the microenvironment significantly influences cancer progression. Here, we report a significant role of the transcription factor FOXM1 in shaping the tumor immune landscape. Single-cell sequencing reveals that tumor-intrinsic FOXM1 creates an immune-suppressive tumor microenvironment by inhibiting expression of stress ligands (including ULBP1) on cancer cells, thereby blocking NKG2D-NKG2DL interactions critical for priming natural killer- and T cell-mediated cytotoxicity of cancer cells. FOXM1 suppresses ULBP1 expression by epigenetically silencing the DNA-sensing protein STING using a DNMT1-UHRF1 complex, which in turn inhibits the unfolded protein response protein CHOP from activating ULBP1. Importantly, cancer patients with higher levels of FOXM1 and DNMT1, and lower levels of STING and ULBP1, have worse survival and are less responsive to immunotherapy. Collectively, our findings provide key insight into how a tumor-intrinsic transcription factor epigenetically shapes the tumor immune microenvironment, with strong implications for refining existing and designing new cancer immunotherapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
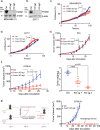





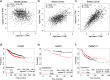
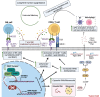
References
-
- Wellenstein, M. D. & Visser, K. E. Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity48, 399–416 (2018). - PubMed
-
- Spranger, S. & Gajewski, T. F. Mechanisms of tumor cell–intrinsic immune evasion. Annu. Rev. Cancer Biol.2, 213–228 (2018).
MeSH terms
Substances
Grants and funding
- T32 GM113896/GM/NIGMS NIH HHS/United States
- R01CA179120/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- T32 GM145432/GM/NIGMS NIH HHS/United States
- T32 CA148724/CA/NCI NIH HHS/United States
- T32 CA279363/CA/NCI NIH HHS/United States
- I01 BX006280/BX/BLRD VA/United States
- P30 CA054174/CA/NCI NIH HHS/United States
- I01 BX005955/BX/BLRD VA/United States
- R01 CA179120/CA/NCI NIH HHS/United States
- R01CA239227/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA239227/CA/NCI NIH HHS/United States
- S10 OD030311/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

