Dissecting cross-population polygenic heterogeneity across respiratory and cardiometabolic diseases
- PMID: 40295474
- PMCID: PMC12037804
- DOI: 10.1038/s41467-025-58149-y
Dissecting cross-population polygenic heterogeneity across respiratory and cardiometabolic diseases
Abstract
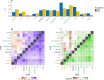
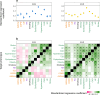
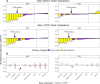
Biological mechanisms underlying multimorbidity remain elusive. To dissect the polygenic heterogeneity of multimorbidity in twelve complex traits across populations, we leveraged biobank resources of genome-wide association studies (GWAS) for 232,987 East Asian individuals (the 1st and 2nd cohorts of BioBank Japan) and 751,051 European individuals (UK Biobank and FinnGen). Cross-trait analyses of respiratory and cardiometabolic diseases, rheumatoid arthritis, and smoking identified negative genetic correlations between respiratory and cardiometabolic diseases in East Asian individuals, opposite from the positive associations in European individuals. Associating genome-wide polygenic risk scores (PRS) with 325 blood metabolome and 2917 proteome biomarkers supported the negative cross-trait genetic correlations in East Asian individuals. Bayesian pathway PRS analysis revealed a negative association between asthma and dyslipidemia in a gene set of peroxisome proliferator-activated receptors. The pathway suggested heterogeneity of cell type specificity in the enrichment analysis of the lung single-cell RNA-sequencing dataset. Our study highlights the heterogeneous pleiotropy of immunometabolic dysfunction in multimorbidity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

