Investigative needle core biopsies support multimodal deep-data generation in glioblastoma
- PMID: 40295505
- PMCID: PMC12037860
- DOI: 10.1038/s41467-025-58452-8
Investigative needle core biopsies support multimodal deep-data generation in glioblastoma
Abstract
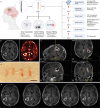
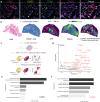
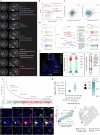
Glioblastoma (GBM) is an aggressive primary brain cancer with few effective therapies. Stereotactic needle biopsies are routinely used for diagnosis; however, the feasibility and utility of investigative biopsies to monitor treatment response remains ill-defined. Here, we demonstrate the depth of data generation possible from routine stereotactic needle core biopsies and perform highly resolved multi-omics analyses, including single-cell RNA sequencing, spatial transcriptomics, metabolomics, proteomics, phosphoproteomics, T-cell clonotype analysis, and MHC Class I immunopeptidomics on standard biopsy tissue obtained intra-operatively. We also examine biopsies taken from different locations and provide a framework for measuring spatial and genomic heterogeneity. Finally, we investigate the utility of stereotactic biopsies as a method for generating patient-derived xenograft (PDX) models. Multimodal dataset integration highlights spatially mapped immune cell-associated metabolic pathways and validates inferred cell-cell ligand-receptor interactions. In conclusion, investigative biopsies provide data-rich insight into disease processes and may be useful in evaluating treatment responses.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: C.B. is a consultant for: Depuy-Synthes, GalecVn Therapeutics, Haystack Oncology, Privo Technologies and Bionaut Labs, and co-founder of OrisDx and Belay Diagnostics. A.B. is on the Scientific Advisory Board in an unpaid capacity for Evren Technologies and is an inventor on the following patents: 62/258,044, 10/413,522, and 63/052,139, as well as the US Provisional Patent Applications No. 63/449,817, and 63/449,823, and the International Patent Application No. PCT/US24/18343 filed by MSKCC. M.H. is on the Data Safety Monitoring board for Paraxel, Advarra; on the Scientific Advisory Board for Bayer, AnHeartTherapeuVcs Inc, and Servier; and speaking engagement with Novartis. D.R. is a consultant for: AnHeart Pharmaceuticals, Aptitude Health, BlueRock Therapeutics LP, CeCaVaGmbH & Co.KG, Chimeric Therapeutics, Elsevier, Enterome, F. Hoffman La-Roche, Genenta Science, Inovio, Insightec, Janssen, Jupiter Life Sciences Consulting, LLC, Kintara, Kiyatec, Johnson & Johnson, Pharma, Lumanity, Menari Stemline, Miltenyi Biomedicine GmbH, Neuvogen, Novocure, Paradigm Medical Communications, Putnam Inizii Associates, LLC, Sumitono Dainippon Pharma Oncology, Servier, Triangle Insights Group, Vivacitas Oncology, Inc., WebMD; and is on the Data Safety Monitoring Board for ImVax, CeCaVaGmbH; Private Investment in AnHeart Therapeutics, Bionaut Lab, and has received research funding via DFCI from Acerta Phamaceuticals, Agenus, Ashvattha Therapeutics, Boehringer Ingelheim, Bristol-Myers Squibb, Corbus Pharma, EMD Serono, Enterome, Epitopoietic Research Corporation, Incyte, Inovio, Insightec, Invios, Merck, Medicenna Therapeutics, Mogling Bio, NeoTx Ltd, Numiera Therapeutics, Sapience Therapeutics, SphereBio, Vaccinex. P.S. is on the Scientific Advisory Board for: Achelois, Akoya Biosciences, Affini-T, Apricity, Asher Bio, BioAtla LLC, Candel Therapeutics, Catalio, C-Reveal Therapeutics, Dragonfly Therapeutics, Earli Inc, Enable Medicine, Glympse, Henlius/Hengenix, Hummingbird, ImaginAb, InterVenn Biosciences, LAVA Therapeutics, Lytix Biopharma, Marker Therapeutics, Matrisome, NTx, Oncolytics, Osteologic, PBM Capital, Phenomic AI, Polaris Pharma, Soley Therapeutics, Spotlight, Trained Therapeutix Discovery, Two Bear Capital, Vironexis, Xilis, Inc. Private Investment: Adaptive Biotechnologies, BioNTech, JSL Health, Sporos, Time Bioventures. N.A. is a consultant for: Bruker; on the Scientific Advisory Board for: National Brain Tumor Society and received research support from Thermo, EMD Serono and iTeos Therapeutics. R.B. was previously on the Scientific Advisory Board for: Scorpion Therapeutics, co-founder of Karyoverse Therapeutics; owns equity in Karyoverse Therapeutics and Takeda Therapeutics. M.C. is a consultant for: Dare Bioscience, Johnson and Johnson; is director at GelMEDIX Inc., and director and founder of Stratagen Bio. K.L.L. has interests not directly related to the study: Equity holder: Travera Inc., Consultant: Travera Inc, Servier, Bristol-Myers Squibb, Integragen, L.E.K. Consulting, Blaze Bioscience; and received research funding to DFCI: via Bristol-Myers Squibb and Eli-Lilly. L.C.-H. is a consultant for Servier. F.M. is a co-founder of and has equity in Harbinger Health, has equity in Zephyr AI, and serves as a consultant for both companies. She is also on the board of directors of Recursion Pharmaceuticals. None of these relationships are directly or indirectly related to the content of this manuscript. The rest of the authors do not have competing interests to declare.
Figures





Update of
-
Investigative needle core biopsies for multi-omics in Glioblastoma.medRxiv [Preprint]. 2023 Dec 31:2023.12.29.23300541. doi: 10.1101/2023.12.29.23300541. medRxiv. 2023. Update in: Nat Commun. 2025 Apr 28;16(1):3957. doi: 10.1038/s41467-025-58452-8. PMID: 38234840 Free PMC article. Updated. Preprint.
References
-
- Weller, M. et al. Glioma. Nat. Rev. Dis. Prim.1, 15017 (2015). - PubMed
-
- Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med.352, 987–996 (2005). - PubMed
-
- Stupp, R. et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol.10, 459–466 (2009). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

