A 6-tsRNA signature for early detection, treatment response monitoring, and prognosis prediction in diffuse large B cell lymphoma
- PMID: 40295511
- PMCID: PMC12037784
- DOI: 10.1038/s41408-025-01267-z
A 6-tsRNA signature for early detection, treatment response monitoring, and prognosis prediction in diffuse large B cell lymphoma
Abstract
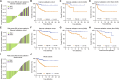
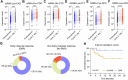
Diffuse large B-cell lymphoma (DLBCL) presents considerable clinical challenges due to its aggressive nature and diverse clinical progression. New molecular biomarkers are urgently needed for outcome prediction. We analyzed blood samples from DLBCL patients and healthy individuals using short, non-coding RNA sequencing. A classifier based on six tsRNAs was developed through random forest and primary component analysis. This classifier, established using Cox proportional hazards modeling with repeated 10-fold cross-validation on an internal cohort of 100 samples analyzed using RT-qPCR, effectively identified high-risk patients with significantly lower overall survival compared to low-risk patients (Hazard ratio: 6.657, 95%CI 2.827-15.68, P = 0.0006). Validation in an external cohort of 160 samples using RT-qPCR confirmed the classifier's robust performance. High-risk status was strongly associated with disease histological subtype, stage, and International Prognostic Index scores. Integration of the classifier into the IPI model enhanced the precision and consistency of prognostic predictions. A dynamic study revealed that patients experiencing a 1.06-fold decrease after one therapy cycle (early molecular response) exhibited better treatment outcomes and prognosis. Furthermore, the 6-tsRNA signature accurately differentiated healthy individuals from DLBCL (AUC 0.882, 95%CI 0.826-0.939). These findings underscore the potential of the identified 6-tsRNA profile as a biomarker for monitoring treatment effectiveness and predicting DLBCL outcomes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
MicroRNAs associated to single drug components of R-CHOP identifies diffuse large B-cell lymphoma patients with poor outcome and adds prognostic value to the international prognostic index.BMC Cancer. 2020 Mar 20;20(1):237. doi: 10.1186/s12885-020-6643-8. BMC Cancer. 2020. PMID: 32192453 Free PMC article.
-
Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma.Mol Cancer. 2017 Jan 19;16(1):16. doi: 10.1186/s12943-017-0580-4. Mol Cancer. 2017. PMID: 28103885 Free PMC article.
-
Transcriptome profiling reveals an integrated mRNA-lncRNA signature with predictive value for long-term survival in diffuse large B-cell lymphoma.Aging (Albany NY). 2020 Nov 18;12(22):23275-23295. doi: 10.18632/aging.104100. Epub 2020 Nov 18. Aging (Albany NY). 2020. PMID: 33221755 Free PMC article.
-
Diffuse large B-cell lymphoma.Pathology. 2018 Jan;50(1):74-87. doi: 10.1016/j.pathol.2017.09.006. Epub 2017 Nov 20. Pathology. 2018. PMID: 29167021 Review.
-
Remaining challenges in predicting patient outcomes for diffuse large B-cell lymphoma.Expert Rev Hematol. 2019 Nov;12(11):959-973. doi: 10.1080/17474086.2019.1660159. Epub 2019 Sep 12. Expert Rev Hematol. 2019. PMID: 31513757 Free PMC article. Review.
References
-
- Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin. 2016;66:443–59. - PubMed
-
- Poletto S, Novo M, Paruzzo L, Frascione PMM, Vitolo U. Treatment strategies for patients with diffuse large B-cell lymphoma. Cancer Treat Rev. 2022;110:102443. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

