Multi-country and intersectoral assessment of cluster congruence between pipelines for genomics surveillance of foodborne pathogens
- PMID: 40295532
- PMCID: PMC12038046
- DOI: 10.1038/s41467-025-59246-8
Multi-country and intersectoral assessment of cluster congruence between pipelines for genomics surveillance of foodborne pathogens
Abstract
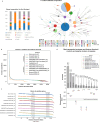
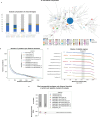
Different laboratories employ different Whole-Genome Sequencing (WGS) pipelines for Food and Waterborne disease (FWD) surveillance, casting doubt on the comparability of their results and hindering optimal communication at intersectoral and international levels. Through a collaborative effort involving eleven European institutes spanning the food, animal, and human health sectors, we aimed to assess the inter-pipeline clustering congruence across all resolution levels and perform an in-depth comparative analysis of cluster composition at outbreak level for four important foodborne pathogens: Listeria monocytogenes, Salmonella enterica, Escherichia coli, and Campylobacter jejuni. We found a general concordance between allele-based pipelines for all species, except for C. jejuni, where the different resolution power of allele-based schemas led to marked discrepancies. Still, we identified non-negligible differences in outbreak detection and demonstrated how a threshold flexibilization favors the detection of similar outbreak signals by different laboratories. These results, together with the observation that different traditional typing groups (e.g., serotypes) exhibit a remarkably different genetic diversity, represent valuable information for future outbreak case-definitions and WGS-based nomenclature design. This study reinforces the need, while demonstrating the feasibility, of conducting continuous pipeline comparability assessments, and opens good perspectives for a smoother international and intersectoral cooperation towards an efficient One Health FWD surveillance.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- WHO. Estimating the burden of foodborne diseases: A practical handbook for countries. https://www.who.int/publications/i/item/9789240012264 (2021).
-
- Struelens, M. J. et al. Real-time genomic surveillance for enhanced control of infectious diseases and antimicrobial resistance. Front. Sci. Ser.2, 1298248 (2024).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

