Differential behavior responses and genetic alteration underpinning exercise effectiveness in stress-susceptible mice
- PMID: 40295575
- PMCID: PMC12038050
- DOI: 10.1038/s41598-025-98630-8
Differential behavior responses and genetic alteration underpinning exercise effectiveness in stress-susceptible mice
Abstract
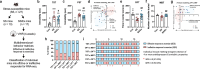
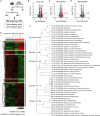
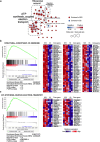
Stress susceptibility varies across individuals, influenced by genetic, molecular, and environmental factors. Hence, approaches for exercise treatment as an antidepressant and anxiolytic intervention must consider individual variability. Examining individual adaptation to exercise provides insights into the biology of such variations. We investigated the efficacy of voluntary wheel running (VWR) exercise as a disease-modifying treatment for stress-susceptible (SS) mice subjected to chronic restraint stress. A multidimensional behavior analysis revealed significant variability in VWR efficacy among individuals; while some mice showed substantial behavior phenotypic improvements (SES), others displayed limited/no benefits (SER). A transcriptomic profiling of the ventral hippocampus, a brain region critical to emotional regulation, revealed molecular signatures that promote adaptive changes by restoring cellular repair, energy availability, and synaptic plasticity in SS mice. SER mice exhibited limited behavior resilience and distinct transcriptomic profiles enriched in structural adaptation without functional resilience and glial cell differentiation marked by astrocyte activation or differentiation. These findings suggest that while VWR can mitigates multiple behavior symptoms in stress-susceptible mice, its effectiveness is modulated by distinct biological mechanisms. We highlight the importance of a multivariate framework for behavior assessment and genetic underpinnings, clarifying the variability in responses to stress and exercise's therapeutic efficacy in stress-related disorders.
Keywords: Anxiety; Depression; Exercise; Resilience; Stress; Susceptibility.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
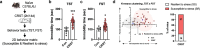



Similar articles
-
Voluntary wheel running during adolescence prevents the increase in ethanol intake induced by social defeat in male mice.Psychopharmacology (Berl). 2025 May;242(5):979-996. doi: 10.1007/s00213-023-06461-0. Epub 2023 Sep 22. Psychopharmacology (Berl). 2025. PMID: 37736785 Free PMC article.
-
Voluntary wheel running promotes resilience to chronic social defeat stress in mice: a role for nucleus accumbens ΔFosB.Neuropsychopharmacology. 2018 Aug;43(9):1934-1942. doi: 10.1038/s41386-018-0103-z. Epub 2018 May 24. Neuropsychopharmacology. 2018. PMID: 29875450 Free PMC article.
-
Lactate is an antidepressant that mediates resilience to stress by modulating the hippocampal levels and activity of histone deacetylases.Neuropsychopharmacology. 2019 May;44(6):1152-1162. doi: 10.1038/s41386-019-0313-z. Epub 2019 Jan 8. Neuropsychopharmacology. 2019. PMID: 30647450 Free PMC article.
-
Voluntary wheel running ameliorates depression-like behaviors and brain blood oxygen level-dependent signals in chronic unpredictable mild stress mice.Behav Brain Res. 2017 Jul 14;330:17-24. doi: 10.1016/j.bbr.2017.05.032. Epub 2017 May 17. Behav Brain Res. 2017. PMID: 28527694
-
Functional kinome profiling reveals brain protein kinase signaling pathways and gene networks altered by acute voluntary exercise in rats.PLoS One. 2025 Apr 15;20(4):e0321596. doi: 10.1371/journal.pone.0321596. eCollection 2025. PLoS One. 2025. PMID: 40233052 Free PMC article.
References
-
- Krishnan, V. et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell131, 391–404 (2007). - PubMed
-
- Ebner, K. & Singewald, N. Individual differences in stress susceptibility and stress inhibitory mechanisms. Curr. Opin. Behav. Sci.14, 54–64 (2017).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

