Incompletely closed HIV-1CH040 envelope glycoproteins resist broadly neutralizing antibodies while mediating efficient HIV-1 entry
- PMID: 40295863
- PMCID: PMC11735869
- DOI: 10.1038/s44298-024-00082-w
Incompletely closed HIV-1CH040 envelope glycoproteins resist broadly neutralizing antibodies while mediating efficient HIV-1 entry
Abstract
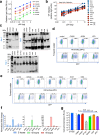
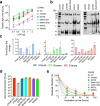
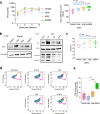
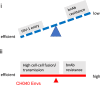
HIV-1 envelope glycoproteins (Envs) mediate viral entry and are sole target of neutralizing antibodies. Thus, HIV-1 Envs must maintain a delicate balance between evading neutralizing antibodies while still preserving viral compatibility to mediate entry into target cells. Here, we studied the viral entry effeciency, fitness, and replication of an incompletely closed, transmitted/founder HIV-1 Envs (CH040), which are highly resistant to most bnAbs. CH040 Envs mediated HIV-1 entry to target cells as efficient as other primary Envs, suggesting that antibody resistance and efficient viral entry can develop independently. Expression of CH040 Envs was comparable to other Envs and most CH040 variants that were rationally engineered to increase bnAb resistance showed no significant decrease in their ability to mediate HIV-1 entry. We detected robust in vitro spread of SHIV CH040 in pig-tailed macaque lymphocytes that was comparable to efficient spread of other SHIVs. Our study provides insights into the relationship between bnAb resistance and efficient HIV-1 entry.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.H. is an inventor on a patent application filed by the University of Minnesota for engineering Env immunogens and the founder of SyntIV LLC. Other authors declare no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

