De novo, high-quality assembly and annotation of the halophyte grass Aeluropus littoralis draft genome and identification of A20/AN1 zinc finger protein family
- PMID: 40295936
- PMCID: PMC12039208
- DOI: 10.1186/s12870-025-06610-x
De novo, high-quality assembly and annotation of the halophyte grass Aeluropus littoralis draft genome and identification of A20/AN1 zinc finger protein family
Abstract
Background: Aeluropus littoralis is considered a valuable natural forage plant for ruminant livestock and is highly tolerant to extreme abiotic stresses, especially salinity, drought, and heat. It is a monocotyledonous halophyte, has salt glands, performs C4-type photosynthesis and has a close genetic relationship with cereal crops. Moreover, previous studies have shown its huge potential as a reservoir of genes and promoters to understand and improve abiotic stress tolerance in crops.
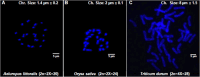
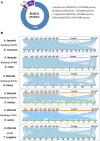
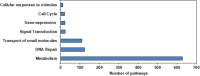
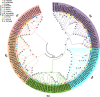
Results: The sequencing and hybrid assembly of the A. littoralis genome (2n = 2X = 20) using short and long reads from the BGISeq-500 and PacBio high-fidelity (HiFi) sequencing platforms, respectively. Using the k-mer analysis method, the haploid genome size of A. littoralis was estimated to be 360 Mb (with a heterozygosity rate of 1.88%). The hybrid assembled genome included 4,078 contigs with a GC content of 44% and covered 348 Mb. The longest contig and the N50 values were 5.1 Mb and 133.77 kb, respectively. The Benchmarking Universal Single Copy Ortholog (BUSCO) value was 91.1%, indicating good integrity of the assembled genome. The discovered repetitive elements accounted for 90.6 Mb, representing 26.03% of the total genome, and included a significant component of transposable elements (11.48%, ~40 Mb). Using a homology-based approach, 35,147 genes were predicted from the genome assembly. We next focused our analysis on the zinc-finger A20/AN1 gene family, a member of which (AlSAP) was previously shown to confer increased tolerance to osmotic and salt stresses when it was over-expressed in tobacco, wheat, and rice. Here, we identified the complete set of members of this family in the Aeluropus littoralis genome, thereby laying the foundation for their future functional analysis in cereal crops. In addition, the expression patterns of four novel genes from this family were analyzed by qPCR.
Conclusion: This resource and our findings will contribute to improve the current understanding of salinity tolerance in halophytes while providing useful genes and allelic variation to improve salinity and drought tolerance in cereals through genetic engineering and gene editing, respectively.
Keywords: Aeluropus littoralis; Abiotic stress tolerance; Halophyte; Next generation sequencing; Zinc-finger A20/AN1 genes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The authors have respected the relevant institutional, national and international guidelines in collecting biological materials for this work. This research contributes to facilitating future studies in comparative genomics, phylogeny, and genomic-assisted breeding program. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Improved drought and salt stress tolerance in transgenic tobacco overexpressing a novel A20/AN1 zinc-finger "AlSAP" gene isolated from the halophyte grass Aeluropus littoralis.Plant Mol Biol. 2010 Jan;72(1-2):171-90. doi: 10.1007/s11103-009-9560-4. Epub 2009 Oct 17. Plant Mol Biol. 2010. PMID: 19838809
-
Genome of extreme halophyte Puccinellia tenuiflora.BMC Genomics. 2020 Apr 19;21(1):311. doi: 10.1186/s12864-020-6727-5. BMC Genomics. 2020. PMID: 32306894 Free PMC article.
-
Promoter of the AlSAP gene from the halophyte grass Aeluropus littoralis directs developmental-regulated, stress-inducible, and organ-specific gene expression in transgenic tobacco.Transgenic Res. 2011 Oct;20(5):1003-18. doi: 10.1007/s11248-010-9474-6. Epub 2010 Dec 28. Transgenic Res. 2011. PMID: 21188636
-
The promoter of the AlSAP gene from the halophyte grass Aeluropus littoralis directs a stress-inducible expression pattern in transgenic rice plants.Plant Cell Rep. 2015 Oct;34(10):1791-806. doi: 10.1007/s00299-015-1825-6. Epub 2015 Jun 30. Plant Cell Rep. 2015. PMID: 26123290
-
Initial Description of the Genome of Aeluropus littoralis, a Halophile Grass.Front Plant Sci. 2022 Jul 11;13:906462. doi: 10.3389/fpls.2022.906462. eCollection 2022. Front Plant Sci. 2022. PMID: 35898222 Free PMC article.
References
-
- Hussain S, Shaukat M, Ashraf M, Zhu C, Jin Q, Zhang J. Salinity stress in arid and semi-arid climates: effects and management in field crops. Clim Change Agric IntechOpen. 2019;13:201–26.
-
- Etikala B, Adimalla N, Madhav S, Somagouni SG, Keshava Kiran Kumar PL. Salinity problems in groundwater and management strategies in arid and semi‐arid regions. Groundwater geochemistry. Wiley; 2021. p. 42–56. 10.1002/9781119709732.ch3.
-
- dos Santos TB, Ribas AF, de Souza SGH, Budzinski IGF, Domingues DS. Physiological responses to drought, salinity, and heat stress in plants: a review. Stresses. 2022;2(1):113–35. 10.3390/stresses2010009.
MeSH terms
Substances
Grants and funding
- Award number 2-17-04-001-0046/The National Plan for Science, Technology and Innovation (MAARIFAH), King Abdul Aziz City for Science and Technology, Kingdom of Saudi Arabia
- Award number 2-17-04-001-0046/The National Plan for Science, Technology and Innovation (MAARIFAH), King Abdul Aziz City for Science and Technology, Kingdom of Saudi Arabia
LinkOut - more resources
Full Text Sources
Miscellaneous

