Stable Plasmodium falciparum merozoite surface protein-1 allelic diversity despite decreasing parasitaemia in children with multiple malaria infections
- PMID: 40296022
- PMCID: PMC12036178
- DOI: 10.1186/s12936-025-05378-7
Stable Plasmodium falciparum merozoite surface protein-1 allelic diversity despite decreasing parasitaemia in children with multiple malaria infections
Abstract
Background: Individuals experiencing recurrent malaria infections encounter a variety of alleles with each new infection. This ongoing allelic diversity influences the development of naturally acquired immunity and it can inform vaccine efficacy. To investigate the diversity and infection variability, Plasmodium falciparum merozoite surface protein 1 (PfMSP1), a crucial protein for parasite invasion and immune response, was assessed in parasites isolated from children in the Junju cohort, Kilifi County, who experienced at least 10 febrile malaria episodes over a span of 5 years.
Methods: Pfmsp1 C-terminal region (Pfmsp119) was genotyped using PCR followed by capillary sequencing in blood samples collected from the children. Sequenced reads were trimmed and aligned to the P. falciparum 3D7 reference genome. Single nucleotide polymorphisms in the Pfmsp119 region were identified from the alignment and grouped to distinct microhaplotypes whose changing frequency over time were examined across the multiple infection episodes. In addition, the variability of infections in the population was assessed using nucleotide and haplotype diversity indices.
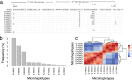
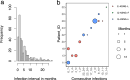
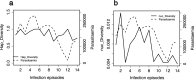
Results: A total of eleven microhaplotypes were observed across all malaria episodes. There were 3 prevalent microhaplotypes, E-KSNG-L, Q-KSNG-L, and Q-KSNG-F in the population. Conversely, microhaplotypes such as Q-KNNG-L, E-KSSR-L, E-KNNG-L, E-KSSG-L, E-TSSR-L (3D7), Q-TSSR-L, E-TSSG-L, and E-KSNG-F were rare and maintained at low frequencies. High allelic replacements were observed, however some individuals experienced consecutive re-infections with the same microhaplotype. Notably, PfMSP119 allelic diversity as measured by haplotype diversity was stable, while nucleotide diversity decreased over time with decreasing parasitemia. Parasite PfMSP119 allelic diversity remained stable over the multiple malaria episodes, despite declining parasitaemia levels. In addition, there are reveal dynamic PfMSP119 allelic replacements across parasite infection episodes.
Conclusions: Allelic diversity was stable over time in individuals, based on this limited polymorphic region and small sample size, suggesting that there are no significant shifts in allele frequencies or replacements due to alleles being maintained under balancing selection. The dominant alleles in the population are those frequently observed in these children with multiple malaria episodes, further suggesting that early exposure to dominant alleles does not shift their frequency in the population or prevent repeat infection with the same alleles in subsequent infections. However, a blood stage merozoite vaccine is likely to require a multi-allelic formulation.
Keywords: Pfmsp1; Diversity; Malaria; Microhaplotype; Parasitaemia.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
The Plasmodium falciparum merozoite surface protein-1 19 KD antibody response in the Peruvian Amazon predominantly targets the non-allele specific, shared sites of this antigen.Malar J. 2010 Jan 4;9:3. doi: 10.1186/1475-2875-9-3. Malar J. 2010. PMID: 20047674 Free PMC article.
-
Effect of malaria transmission reduction by insecticide-treated bed nets (ITNs) on the genetic diversity of Plasmodium falciparum merozoite surface protein (MSP-1) and circumsporozoite (CSP) in western Kenya.Malar J. 2013 Aug 27;12:295. doi: 10.1186/1475-2875-12-295. Malar J. 2013. PMID: 23978002 Free PMC article. Clinical Trial.
-
Plasmodium falciparum msp1 and msp2 genetic diversity and allele frequencies in parasites isolated from symptomatic malaria patients in Bobo-Dioulasso, Burkina Faso.Parasit Vectors. 2018 May 30;11(1):323. doi: 10.1186/s13071-018-2895-4. Parasit Vectors. 2018. PMID: 29843783 Free PMC article.
-
Population diversity and antibody selective pressure to Plasmodium falciparum MSP1 block2 locus in an African malaria-endemic setting.BMC Microbiol. 2009 Oct 15;9:219. doi: 10.1186/1471-2180-9-219. BMC Microbiol. 2009. PMID: 19832989 Free PMC article.
-
Genetic diversity of Plasmodium falciparum and genetic profile in children affected by uncomplicated malaria in Cameroon.Malar J. 2020 Mar 18;19(1):115. doi: 10.1186/s12936-020-03161-4. Malar J. 2020. PMID: 32188442 Free PMC article. Clinical Trial.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

