A genome-wide One Health study of Klebsiella pneumoniae in Norway reveals overlapping populations but few recent transmission events across reservoirs
- PMID: 40296028
- PMCID: PMC12039103
- DOI: 10.1186/s13073-025-01466-0
A genome-wide One Health study of Klebsiella pneumoniae in Norway reveals overlapping populations but few recent transmission events across reservoirs
Abstract
Background: Members of the Klebsiella pneumoniae species complex (KpSC) are opportunistic pathogens that cause severe and difficult-to-treat infections. KpSC are common in non-human niches, but the clinical relevance of these populations is disputed.
Methods: In this study, we analysed 3255 whole-genome sequenced isolates from human, animal and marine sources collected in Norway between 2001 and 2020. We used population genomics in a One Health context to assess the diversity of strains, genes and other clinically relevant genetic features within and between sources. We further explored niche-enriched traits using genome-wide association studies and investigated evidence of spillover and connectivity across the KpSC populations from the three niches.
Results: We found that the KpSC populations in different niches were distinct but overlapping. Overall, there was high genetic diversity both between and within sources, with nearly half (49%) of the genes in the accessory genome overlapping the ecological niches. Further, several sublineages (SLs) including SL17, SL35, SL37, SL45, SL107 and SL3010 were common across sources. There were few niche-enriched traits, except for aerobactin-encoding plasmids and the bacteriocin colicin a, which were associated with KpSC from animal sources. Human infection isolates showed the greatest connectivity with each other, followed by isolates from human carriage, pigs, and bivalves. Nearly 5% of human infection isolates had close relatives (≤22 substitutions) amongst animal and marine isolates, despite temporally and geographically distant sampling of these sources. There were limited but notable recent spillover events, including the movement of plasmids encoding the virulence locus iuc3 between pigs and humans.
Conclusions: Our large One Health genomic study highlights that human-to-human transmission of KpSC is more common than transmission between ecological niches. Still, spillover of clinically relevant strains and genetic features between human and non-human sources does occur and should not be overlooked. Infection prevention measures are essential to limit transmission within human clinical settings and reduce infections. However, preventing transmission that leads to colonisation, e.g. from direct contact with animals or via the food chain, could also play an important role in reducing the KpSC disease burden.
Keywords: Klebsiella pneumoniae species complex; AMR; Ecology; GWAS; Genomics; One Health; Transmission; Zoonotic transmission.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study collecting the human faecal carriage isolates was approved by the Regional Committee for Medical and Health Research Ethics, North Norway (REC North reference: 2016/1788). The two studies collecting human infection isolates were approved by the Regional Committee for Medical and Health Research Ethics, Western Norway (REC West reference 2017/1185 and 2016/1093). No additional approvals were required for using the human faecal carriage and infection KpSC genomes for this study as we did not collect any additional samples or data. The three studies are published 21,22,25 and the genomes included in the studies are publicly available (in BioProjects: PRJEB42350, PRJEB48268 and PRJEB27256). The research conformed to the principles of the Helsinki Declaration. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
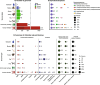

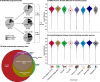


References
-
- Palma F, Hennart M, Jolley KA, et al. Bacterial strain nomenclature in the genomic era: Life Identification Numbers using a gene-by-gene approach. 2024. Preprint at bioRxiv. 10.1101/2024.03.11.584534
MeSH terms
Grants and funding
- F-12508/Helse Vest Regionalt Helseføretak
- 912119/Helse Vest Regionalt Helseføretak
- 912037/Helse Vest Regionalt Helseføretak
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- TMF2019TMT03/Trond Mohn stiftelse
- HNF1415-18/Helse Nord RHF
- HNF1415-18/Helse Nord RHF
LinkOut - more resources
Full Text Sources

