DNA methylation signatures of severe RSV infection in infants: evidence from non-invasive saliva samples
- PMID: 40296166
- PMCID: PMC12036262
- DOI: 10.1186/s13072-025-00587-5
DNA methylation signatures of severe RSV infection in infants: evidence from non-invasive saliva samples
Abstract
Background: Respiratory syncytial virus (RSV) poses significant morbidity and mortality risks in childhood, particularly for previously healthy infants admitted to hospitals lacking predisposing risk factors for severe disease. This study aimed to investigate the role of the host epigenome in RSV infection severity using non-invasive buccal swabs from sixteen hospitalized infants admitted to the hospital for RSV infection. Eight patients had severe symptoms, and eight had mild to moderate symptoms. For DNA methylation analyses, the Illumina EPIC BeadChip was used with DNA isolated from saliva samples. To evaluate the basal DNA methylation level of the identified biomarkers a cohort of healthy control children was used. Furthermore, DNA methylation levels of candidate genes were confirmed by pyrosequencing in both the discovery and validation cohorts of patients with mild to moderate symptoms.
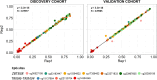
Results: A panel of differentially methylated positions (DMPs) distinguishing severe from mild to moderate symptoms in infants was identified. DMPs were determined using a threshold of an adjusted P-value (false discovery rate, FDR) < 0.01 and an absolute difference in DNA methylation (delta beta) > 0.10. Differentially methylated regions (DMRs) were identified in the ZBTB38 (implicated in asthma and pulmonary disease) and the TRIM6-TRM34 gene region (associated with viral infections). The differential DNA methylation of these genes was validated in an independent replication cohort. A weighted correlation network analysis emphasized the pivotal role of a module with RAB11FIP5 as the hub gene, known for its critical function in regulating viral infections.
Conclusions: Oral mucosa methylation may play a role in determining the severity of RSV disease in infants.
Keywords: Buccal swab; DNA methylation; Epigenetic biomarkers; Respiratory syncytial virus; Severity.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study involving human participants was reviewed and approved by the Ethics Committee of Clinical Investigation of Galicia (CEIC 2016/484). Written informed consent to participate in this study was provided by the participant’s legal guardian/next of kin. This project is conducted following the ICH Harmonized Tripartite Guidelines for Good Clinical Practice, with the current national regulations (Law 14/2007 on Biomedical Research), and with the ethical principles established in the Declaration of Helsinki. The confidentiality of the data of the study participants will be guaranteed, ensuring compliance with Organic Law 3/2018, of December 5, on the Protection of Personal Data and guarantee of digital rights. Consent for publication: Not Applicable. Competing interests: Ana Isabel Dacosta Urbieta has participated in clinical trials for RSV vaccines and monoclonals with all honoraria paid to the institution. Irene Rivero Calle has received honoraria from GSK, Pfizer, Sanofi, and MSD for taking part in advisory boards and expert meetings and acting as a speaker in congresses outside the scope of the submitted work. IRC has also acted as sub-investigator in randomized controlled trials of Abbot, Astrazeneca, Enanta, Gilead, GlaxoSmithKline, Janssen, Medimmune, Merck, Moderna, MSD, Novavax, Pfizer, Reviral, Roche, Sanofi Pasteur, and Seqirus.
Figures








References
-
- Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. The Lancet. 2017;390(10098):946–58. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- PI24/00771/Instituto de Salud Carlos III
- CP23/00080/Instituto de Salud Carlos III
- ReSVinext: PI16/01569; Enterogen: PI19/01090 [F.M.T.]; OMI-COVI-VAC: PI22/00406/Instituto de Salud Carlos III
- TRINEO: PI22/00162; DIAVIR: DTS19/00049; Resvi-Omics: PI19/01039/Instituto de Salud Carlos III
- IN677D 2024/06/Axencia Galega de Innovación
- GEN-COVID: IN845D 2020/23 and IIN607A2021/05/Axencia Galega de Innovación
- IN607B 2020/08, and IN607A 2023/02/Axencia Galega de Innovación
- PID2022-142156OB-I00/Spanish Ministry of Science and Innovation (MCIN)/Spanish Research Agency (AEI)
- Respisal PRIST-VAL/Agencia Gallega de Conocimiento en Salud
- BI-BACVIR and CovidPhy/Agencia Gallega de Conocimiento en Salud
- CB21/06/00103/Centro de Investigación Biomédica en Red de Epidemiología y Salud Pública
- CB21/06/00103/Centro de Investigación Biomédica en Red de Epidemiología y Salud Pública
LinkOut - more resources
Full Text Sources
Medical

