Inhibition of ALKBH5 demethylase of m6A pathway potentiates HIV-1 reactivation from latency
- PMID: 40296171
- PMCID: PMC12039216
- DOI: 10.1186/s12985-025-02744-4
Inhibition of ALKBH5 demethylase of m6A pathway potentiates HIV-1 reactivation from latency
Abstract
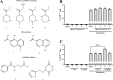
Background: Current latency-reversing agents (LRAs) employed in the "shock-and-kill" strategy primarily focus on relieving epigenetic and transcriptional blocks to reactivate the latent HIV-1. However, their clinical efficacy is limited, partly due to their inability to fully reverse latency and the lack of LRAs specifically targeting post-transcriptional mechanisms. N6-methyladenosine (m6A) modification in HIV-1 RNA is emerging as an important post-transcriptional regulator of HIV-1 gene expression, yet its role in latency and reactivation remains largely unrecognized. Here, we explored the potential of small chemical compounds targeting the m6A pathway, specifically investigating the inhibition of ALKBH5 and its effect on latent HIV-1 reactivation mediated by the LRA romidepsin.
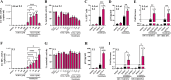
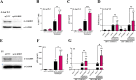
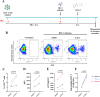
Methods: We used four in vitro cellular models of latency, primary model of CD4+ T cells HIV-1 infection and ex vivo cultures of CD8+-depleted PMBCs from ART-treated HIV+ patients. We measured latent viral reactivation by evaluating the expression of reporter protein GFP by flow cytometry, viral production by CA-p24 ELISA, and viral transcripts by RT-qPCR. CRISPR/Cas9 method was used to deplete ALKBH5. MeRIP and immuno-RNA FISH were used to address the m6A methylation levels on HIV-1 RNA upon ALKBH5 inhibition.
Results: We showed that ALKBH5 inhibitor 3 (ALKi-3) potentiated romidepsin-mediated viral reactivation in in vitro models of latency, primary model of CD4+ T cells infected with HIV-1 as well as in ex vivo cultures of CD8+-depleted PBMCs from ART-treated HIV+ patients. CRISPR/Cas9-mediated depletion of ALKBH5 mimicked the effects of ALKi-3. ALKi-3 increased levels of m6A-methylated HIV-1 RNA as shown by meRIP and immuno-RNA FISH.
Conclusion: Our study provides a proof-of-concept for the modulation of the m6A pathway in enhancing HIV-1 reactivation. This approach represents a promising adjunct to existing reactivation protocols and provides a concept of "dual-kick", aiming to target transcriptional and post-transcriptional steps in HIV-1 reactivation from latency.
Keywords: ALKBH5; Dual-kick; Epitranscriptomics; HIV-1; Latency; Post-transcriptional mechanisms; Shock-and-kill; m6A.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical approval was granted by the Human Subject Ethics Committees of the Saint-Pierre Hospital (Brussels, Belgium). All individuals enrolled in the study provided written informed consent for donating blood. Consent for publication: Not applicable. Conflict of interest: A.O.P. received a research grant from Gilead Sciences Research Program. C.V.L. received a research grant from ViiV Healthcare. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The other authors declare no competing interests. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Tannic acid reactivates HIV-1 latency by mediating CBX4 degradation.J Virol. 2025 Jan 31;99(1):e0117324. doi: 10.1128/jvi.01173-24. Epub 2024 Dec 18. J Virol. 2025. PMID: 39692477 Free PMC article.
-
Integrator complex subunit 12 knockout overcomes a transcriptional block to HIV latency reversal.Elife. 2025 Apr 10;13:RP103064. doi: 10.7554/eLife.103064. Elife. 2025. PMID: 40207620 Free PMC article.
-
BET degraders reveal BRD4 disruption of 7SK and P-TEFb is critical for effective reactivation of latent HIV in CD4+ T-cells.J Virol. 2025 Apr 15;99(4):e0177724. doi: 10.1128/jvi.01777-24. Epub 2025 Mar 11. J Virol. 2025. PMID: 40067013 Free PMC article.
-
Structured treatment interruptions (STI) in chronic unsuppressed HIV infection in adults.Cochrane Database Syst Rev. 2006 Jul 19;2006(3):CD006148. doi: 10.1002/14651858.CD006148. Cochrane Database Syst Rev. 2006. PMID: 16856117 Free PMC article.
-
Optimisation of antiretroviral therapy in HIV-infected children under 3 years of age.Cochrane Database Syst Rev. 2014 May 22;2014(5):CD004772. doi: 10.1002/14651858.CD004772.pub4. Cochrane Database Syst Rev. 2014. PMID: 24852077 Free PMC article.
References
-
- McMyn NF, Varriale J, Fray EJ, Zitzmann C, MacLeod H, Lai J, Singhal A, Moskovljevic M, Garcia MA, Lopez BM, Hariharan V, Rhodehouse K, Lynn K, Tebas P, Mounzer K, Montaner LJ, Benko E, Kovacs C, Hoh R, Simonetti FR, Laird GM, Deeks SG, Ribeiro RM, Perelson AS, Siliciano RF, Siliciano JM. The latent reservoir of inducible, infectious HIV-1 does not decrease despite decades of antiretroviral therapy. J Clin Invest. 2023;133:e171554. - PMC - PubMed
-
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4 + T cells. Nat Med. 2003;9:727–8. - PubMed
-
- Wong JK, Hezareh M, Günthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Recovery of Replication-Competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

