NPAS4 Depletion in POMC Neurons Protects From Obesity and Alters the Feeding-regulated Transcriptome in Male Mice
- PMID: 40296822
- PMCID: PMC12123070
- DOI: 10.1210/endocr/bqaf083
NPAS4 Depletion in POMC Neurons Protects From Obesity and Alters the Feeding-regulated Transcriptome in Male Mice
Abstract
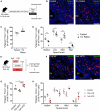
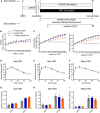
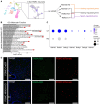
Immediate early genes (IEGs), such as neuronal PAS domain protein 4 (Npas4), are induced as part of the response to environmental stimuli. In the arcuate nucleus (ARC), proopiomelanocortin (POMC) neurons are critical in detecting peripheral signals to regulate food intake. To date, Npas4 has not been studied in the context of regulating food intake, and its sites of action in the ARC are unknown. We found that Npas4 was induced in POMC neurons by refeeding, oral glucose, and a high-fat diet (HFD). In order to explore the role of NPAS4 in POMC neurons, a conditional knockout approach was used. Male mice with Npas4 knockout in POMC neurons showed significantly reduced body weight starting at 10 weeks of HFD, which was due to decreased food intake. Single-cell RNA sequencing on ARC cells demonstrated that POMC neurons of knockout mice showed an enhanced refeeding-induced transcriptional response, dysregulated IEG expression in response to refeeding, and reduced expression of genes encoding gamma-aminobutyric acid (GABA)-A receptor subunits. Cell-to-cell communication analysis revealed that POMC neurons of knockout mice lost inhibitory GABAergic signaling inputs and gained excitatory glutamatergic signaling inputs. Taken together, these results suggest that Npas4 tempers the activity of POMC neurons and loss of Npas4 causes impairments in nutrient intake sensing. Mechanistically, this results from reduced expression of inhibitory GABA-A receptors and an overall increase in the feeding-induced POMC neuron transcriptional response. In conclusion, we report a role for the transcription factor Npas4 in POMC neurons of the ARC and demonstrate its importance in controlling feeding behavior in states of overnutrition.
Keywords: POMC; arcuate nucleus; body weight; immediate early genes; obesity.
© The Author(s) 2025. Published by Oxford University Press on behalf of the Endocrine Society.
Figures




References
-
- Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4(4):477‐485. - PubMed
-
- Abraham WC, Dragunow M, Tate WP. The role of immediate early genes in the stabilization of long-term potentiation. Mol Neurobiol. 1991;5(2–4):297‐314. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

