This is a preprint.
Go Figure: Transparency in neuroscience images preserves context and clarifies interpretation
- PMID: 40297230
- PMCID: PMC12036441
Go Figure: Transparency in neuroscience images preserves context and clarifies interpretation
Abstract
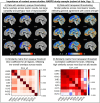
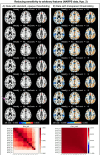
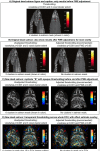
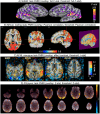
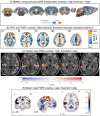
Visualizations are vital for communicating scientific results. Historically, neuroimaging figures have only depicted regions that surpass a given statistical threshold. This practice substantially biases interpretation of the results and subsequent meta-analyses, particularly towards non-reproducibility. Here we advocate for a "transparent thresholding" approach that not only highlights statistically significant regions but also includes subthreshold locations, which provide key experimental context. This balances the dual needs of distilling modeling results and enabling informed interpretations for modern neuroimaging. We present four examples that demonstrate the many benefits of transparent thresholding, including: removing ambiguity, decreasing hypersensitivity to non-physiological features, catching potential artifacts, improving cross-study comparisons, reducing non-reproducibility biases, and clarifying interpretations. We also demonstrate the many software packages that implement transparent thresholding, several of which were added or streamlined recently as part of this work. A point-counterpoint discussion addresses issues with thresholding raised in real conversations with researchers in the field. We hope that by showing how transparent thresholding can drastically improve the interpretation (and reproducibility) of neuroimaging findings, more researchers will adopt this method.
Figures


References
-
- Ad-Dab’bagh Y, Einarson D, Lyttelton O, Muehlboeck J-S, Mok K, Ivanov O, Vincent RD, Lepage C, Lerch J, Fombonne E, Evans AC (2006). The CIVET image-processing environment: A fully automated comprehensive pipeline for anatomical neuroimaging research. Proc. OHBM-2006. http://www.bic.mni.mcgill.ca/users/yaddab/Yasser-HBM2006-Poster.pdf
-
- Amrhein V, Greenland S, McShane B (2019). Scientists rise up against statistical significance. Nature 567:305–307 - PubMed
-
- Anscombe FJ (1973). Graphs in Statistical Analysis. The American Statistician 27(1):17–21
-
- Bacchetti P (2013). Small sample size is not the real problem. Nat Rev Neuroscience 14, 585. - PubMed
Publication types
Grants and funding
- U19 AG073585/AG/NIA NIH HHS/United States
- R01 MH123610/MH/NIMH NIH HHS/United States
- R03 HD113915/HD/NICHD NIH HHS/United States
- R21 HD108587/HD/NICHD NIH HHS/United States
- R01 MH071589/MH/NIMH NIH HHS/United States
- R01 MH124045/MH/NIMH NIH HHS/United States
- P50 MH109429/MH/NIMH NIH HHS/United States
- RF1 MH133701/MH/NIMH NIH HHS/United States
- R01 MH111439/MH/NIMH NIH HHS/United States
- P50 DC014664/DC/NIDCD NIH HHS/United States
- R01 NS109498/NS/NINDS NIH HHS/United States
- R01 AG083919/AG/NIA NIH HHS/United States
- P41 EB019936/EB/NIBIB NIH HHS/United States
- R01 MH096906/MH/NIMH NIH HHS/United States
- R01 EB026859/EB/NIBIB NIH HHS/United States
- R01 DA048993/DA/NIDA NIH HHS/United States
- RF1 MH117040/MH/NIMH NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
