This is a preprint.
Neural Encoding and Decoding at Scale
- PMID: 40297235
- PMCID: PMC12036447
Neural Encoding and Decoding at Scale
Abstract
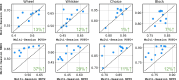
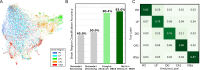
Recent work has demonstrated that large-scale, multi-animal models are powerful tools for characterizing the relationship between neural activity and behavior. Current large-scale approaches, however, focus exclusively on either predicting neural activity from behavior (encoding) or predicting behavior from neural activity (decoding), limiting their ability to capture the bidirectional relationship between neural activity and behavior. To bridge this gap, we introduce a multimodal, multi-task model that enables simultaneous Neural Encoding and Decoding at Scale (NEDS). Central to our approach is a novel multi-task-masking strategy, which alternates between neural, behavioral, within-modality, and cross-modality masking. We pretrain our method on the International Brain Laboratory (IBL) repeated site dataset, which includes recordings from 83 animals performing the same visual decision-making task. In comparison to other large-scale models, we demonstrate that NEDS achieves state-of-the-art performance for both encoding and decoding when pretrained on multi-animal data and then fine-tuned on new animals. Surprisingly, NEDS's learned embeddings exhibit emergent properties: even without explicit training, they are highly predictive of the brain regions in each recording. Altogether, our approach is a step towards a foundation model of the brain that enables seamless translation between neural activity and behavior. Project page and code: https://ibl-neds.github.io/.
Figures


References
-
- Achiam J., Adler S., Agarwal S., Ahmad L., Akkaya I., Aleman F. L., Almeida D., Altenschmidt J., Altman S., Anadkat S., et al. Gpt-4 technical report. arXiv preprint arXiv:2303.08774, 2023.
-
- Azabou M., Arora V., Ganesh V., Mao X., Nachimuthu S., Mendelson M., Richards B., Perich M., Lajoie G., and Dyer E. A unified, scalable framework for neural population decoding. Advances in Neural Information Processing Systems, 36, 2024.
-
- Azabou M., Pan K. X., Arora V., Knight I. J., Dyer E. L., and Richards B. A. Multi-session, multi-task neural decoding from distinct cell-types and brain regions. In The Thirteenth International Conference on Learning Representations, 2025. URL https://openreview.net/forum?id=IuU0wcO0mo.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
