This is a preprint.
REASONING BEYOND ACCURACY: EXPERT EVALUATION OF LARGE LANGUAGE MODELS IN DIAGNOSTIC PATHOLOGY
- PMID: 40297448
- PMCID: PMC12036407
- DOI: 10.1101/2025.04.11.25325686
REASONING BEYOND ACCURACY: EXPERT EVALUATION OF LARGE LANGUAGE MODELS IN DIAGNOSTIC PATHOLOGY
Abstract
Background: Diagnostic pathology depends on complex, structured reasoning to interpret clinical, histologic, and molecular data. Replicating this cognitive process algorithmically remains a significant challenge. As large language models (LLMs) gain traction in medicine, it is critical to determine whether they have clinical utility by providing reasoning in highly specialized domains such as pathology.
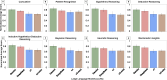
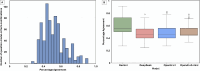
Methods: We evaluated the performance of four reasoning LLMs (OpenAI o1, OpenAI o3-mini, Gemini 2.0 Flash Thinking Experimental, and DeepSeek-R1 671B) on 15 board-style open-ended pathology questions. Responses were independently reviewed by 11 pathologists using a structured framework that assessed language quality (accuracy, relevance, coherence, depth, and conciseness) and seven diagnostic reasoning strategies. Scores were normalized and aggregated for analysis. We also evaluated inter-observer agreement to assess scoring consistency. Model comparisons were conducted using one-way ANOVA and Tukey's Honestly Significant Difference (HSD) test.
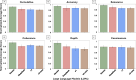
Results: Gemini and DeepSeek significantly outperformed OpenAI o1 and OpenAI o3-mini in overall reasoning quality (p < 0.05), particularly in analytical depth and coherence. While all models achieved comparable accuracy, only Gemini and DeepSeek consistently applied expert-like reasoning strategies, including algorithmic, inductive, and Bayesian approaches. Performance varied by reasoning type: models performed best in algorithmic and deductive reasoning and poorest in heuristic and pattern recognition. Inter-observer agreement was highest for Gemini (p < 0.05), indicating greater consistency and interpretability. Models with more in-depth reasoning (Gemini and DeepSeek) were generally less concise.
Conclusion: Advanced LLMs such as Gemini and DeepSeek can approximate aspects of expert-level diagnostic reasoning in pathology, particularly in algorithmic and structured approaches. However, limitations persist in contextual reasoning, heuristic decision-making, and consistency across questions. Addressing these gaps, along with trade-offs between depth and conciseness, will be essential for the safe and effective integration of AI tools into clinical pathology workflows.
Keywords: AI Evaluation; Clinical Reasoning; Generative AI; Pathology; Reasoning Large Language Models.
Figures


Similar articles
-
DeepSeek-R1 outperforms Gemini 2.0 Pro, OpenAI o1, and o3-mini in bilingual complex ophthalmology reasoning.Adv Ophthalmol Pract Res. 2025 May 9;5(3):189-195. doi: 10.1016/j.aopr.2025.05.001. eCollection 2025 Aug-Sep. Adv Ophthalmol Pract Res. 2025. PMID: 40678192 Free PMC article.
-
A comparative analysis of DeepSeek R1, DeepSeek-R1-Lite, OpenAi o1 Pro, and Grok 3 performance on ophthalmology board-style questions.Sci Rep. 2025 Jul 2;15(1):23101. doi: 10.1038/s41598-025-08601-2. Sci Rep. 2025. PMID: 40595291 Free PMC article.
-
DeepSeek in Healthcare: Revealing Opportunities and Steering Challenges of a New Open-Source Artificial Intelligence Frontier.Cureus. 2025 Feb 18;17(2):e79221. doi: 10.7759/cureus.79221. eCollection 2025 Feb. Cureus. 2025. PMID: 39974299 Free PMC article.
-
Utilizing large language models for gastroenterology research: a conceptual framework.Therap Adv Gastroenterol. 2025 Apr 1;18:17562848251328577. doi: 10.1177/17562848251328577. eCollection 2025. Therap Adv Gastroenterol. 2025. PMID: 40171241 Free PMC article. Review.
-
Challenges and barriers of using large language models (LLM) such as ChatGPT for diagnostic medicine with a focus on digital pathology - a recent scoping review.Diagn Pathol. 2024 Feb 27;19(1):43. doi: 10.1186/s13000-024-01464-7. Diagn Pathol. 2024. PMID: 38414074 Free PMC article.
References
-
- Pena Gil Patrus and Andrade-Filho Josede Souza. How does a pathologist make a diagnosis? Archives of Pathology and Laboratory Medicine, 133(1):124–132, January 2009. - PubMed
-
- Elemento Olivier, Khozin Sean, and Sternberg Cora N. The use of artificial intelligence for cancer therapeutic decision-making. NEJM AI, page AIra2401164, 2025.
-
- Tripathi Aakash, Waqas Asim, Venkatesan Kavya, Ullah Ehsan, Bui Marilyn, and Rasool Ghulam. 1391 ai-driven extraction of key clinical data from pathology reports to enhance cancer registries. Laboratory Investigation, 105(3), 2025.
-
- Wu Jiageng, Liu Xiaocong, Li Minghui, Li Wanxin, Su Zichang, Lin Shixu, Garay Lucas, Zhang Zhiyun, Zhang Yujie, Zeng Qingcheng, et al. Clinical text datasets for medical artificial intelligence and large language models—a systematic review. NEJM AI, 1(6):AIra2400012, 2024.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
